An active role for the ribosome in determining the fate of oxidized mRNA
- PMID: 25456128
- PMCID: PMC4254665
- DOI: 10.1016/j.celrep.2014.10.042
An active role for the ribosome in determining the fate of oxidized mRNA
Abstract
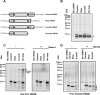
Chemical damage to RNA affects its functional properties and thus may pose a significant hurdle to the translational apparatus; however, the effects of damaged mRNA on the speed and accuracy of the decoding process and their interplay with quality-control processes are not known. Here, we systematically explore the effects of oxidative damage on the decoding process using a well-defined bacterial in vitro translation system. We find that the oxidative lesion 8-oxoguanosine (8-oxoG) reduces the rate of peptide-bond formation by more than three orders of magnitude independent of its position within the codon. Interestingly, 8-oxoG had little effect on the fidelity of the selection process, suggesting that the modification stalls the translational machinery. Consistent with these findings, 8-oxoG mRNAs were observed to accumulate and associate with polyribosomes in yeast strains in which no-go decay is compromised. Our data provide compelling evidence that mRNA-surveillance mechanisms have evolved to cope with damaged mRNA.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

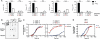

References
-
- Barciszewski J, Barciszewska MZ, Siboska G, Rattan SI, Clark BF. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol. Biol. Rep. 1999;26:231–238. - PubMed
-
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. - PubMed
-
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

