The RNA-editing enzyme ADAR1 controls innate immune responses to RNA
- PMID: 25456137
- PMCID: PMC4542304
- DOI: 10.1016/j.celrep.2014.10.041
The RNA-editing enzyme ADAR1 controls innate immune responses to RNA
Abstract
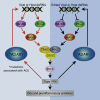
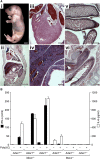
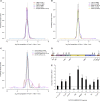
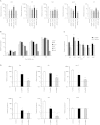
The ADAR RNA-editing enzymes deaminate adenosine bases to inosines in cellular RNAs. Aberrant interferon expression occurs in patients in whom ADAR1 mutations cause Aicardi-Goutières syndrome (AGS) or dystonia arising from striatal neurodegeneration. Adar1 mutant mouse embryos show aberrant interferon induction and die by embryonic day E12.5. We demonstrate that Adar1 embryonic lethality is rescued to live birth in Adar1; Mavs double mutants in which the antiviral interferon induction response to cytoplasmic double-stranded RNA (dsRNA) is prevented. Aberrant immune responses in Adar1 mutant mouse embryo fibroblasts are dramatically reduced by restoring the expression of editing-active cytoplasmic ADARs. We propose that inosine in cellular RNA inhibits antiviral inflammatory and interferon responses by altering RLR interactions. Transfecting dsRNA oligonucleotides containing inosine-uracil base pairs into Adar1 mutant mouse embryo fibroblasts reduces the aberrant innate immune response. ADAR1 mutations causing AGS affect the activity of the interferon-inducible cytoplasmic isoform more severely than the nuclear isoform.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Desterro J.M., Keegan L.P., Lafarga M., Berciano M.T., O’Connell M., Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. - PubMed
-
- Dittmer D., Pati S., Zambetti G., Chu S., Teresky A.K., Moore M., Finlay C., Levine A.J. Gain of function mutations in p53. Nat. Genet. 1993;4:42–46. - PubMed
-
- Funabiki M., Kato H., Miyachi Y., Toki H., Motegi H., Inoue M., Minowa O., Yoshida A., Deguchi K., Sato H. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
Grants and funding
- MC_UU_12021/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127580973/MRC_/Medical Research Council/United Kingdom
- MC_U137761446/MRC_/Medical Research Council/United Kingdom
- MC_U127584490/MRC_/Medical Research Council/United Kingdom
- MC_PC_U127597124/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

