Structural characterization of a flexible two-domain protein in solution using small angle X-ray scattering and NMR data
- PMID: 25456817
- PMCID: PMC5046226
- DOI: 10.1016/j.str.2014.09.013
Structural characterization of a flexible two-domain protein in solution using small angle X-ray scattering and NMR data
Abstract
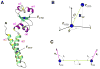
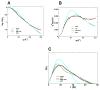
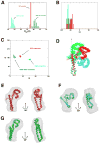
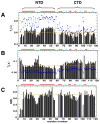
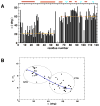
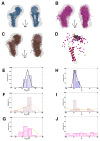
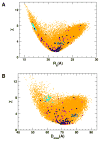
Multidomain proteins in which individual domains are connected by linkers often possess inherent interdomain flexibility that significantly complicates their structural characterization in solution using either nuclear magnetic resonance (NMR) spectroscopy or small-angle X-ray scattering (SAXS) alone. Here, we report a protocol for joint refinement of flexible multidomain protein structures against NMR distance and angular restraints, residual dipolar couplings, and SAXS data. The protocol is based on the ensemble optimization method principle (Bernadó et al., 2007) and is compared with different refinement strategies for the structural characterization of the flexible two-domain protein sf3636 from Shigella flexneri 2a. The results of our refinement suggest the existence of a dominant population of configurational states in solution possessing an overall elongated shape and restricted relative twisting of the two domains.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Almond A, Axeisen JB. Physical Interpretation of Residual Dipolar couplings in neutral Aligned Media. J Am Chem Soc. 2002;124:9986–9987. - PubMed
-
- Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. - PubMed
-
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using Small-Angle X-ray Scattering. J Am Chem Soc. 2007;129:5656–5664. - PubMed
-
- Bertini I, Ferella L, Luchinat C, Parigi G, Petoukhov MV, Ravera E, Rosato A, Svergun DI. MaxOcc: a web portal for maximum occurrence analysis. J Biomol NMR. 2012;53:271–280. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

