PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos
- PMID: 25457166
- PMCID: PMC4250265
- DOI: 10.1016/j.molcel.2014.10.003
PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos
Abstract
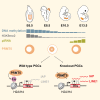
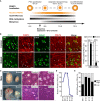
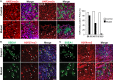
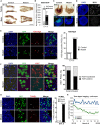
Primordial germ cells (PGCs) and preimplantation embryos undergo epigenetic reprogramming, which includes comprehensive DNA demethylation. We found that PRMT5, an arginine methyltransferase, translocates from the cytoplasm to the nucleus during this process. Here we show that conditional loss of PRMT5 in early PGCs causes complete male and female sterility, preceded by the upregulation of LINE1 and IAP transposons as well as activation of a DNA damage response. Similarly, loss of maternal-zygotic PRMT5 also leads to IAP upregulation. PRMT5 is necessary for the repressive H2A/H4R3me2s chromatin modification on LINE1 and IAP transposons in PGCs, directly implicating this modification in transposon silencing during DNA hypomethylation. PRMT5 translocates back to the cytoplasm subsequently, to participate in the previously described PIWI-interacting RNA (piRNA) pathway that promotes transposon silencing via de novo DNA remethylation. Thus, PRMT5 is directly involved in genome defense during preimplantation development and in PGCs at the time of global DNA demethylation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
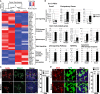
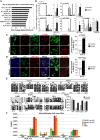

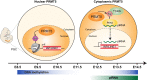
Comment in
-
Prmt5: a guardian of the germline protects future generations.EMBO J. 2015 Mar 12;34(6):689-90. doi: 10.15252/embj.201591054. Epub 2015 Feb 16. EMBO J. 2015. PMID: 25687507 Free PMC article.
References
-
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006;8:623–630. - PubMed
-
- Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H., Forné T., Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

