Rescuing dicer defects via inhibition of an anti-dicing nuclease
- PMID: 25457613
- PMCID: PMC4303555
- DOI: 10.1016/j.celrep.2014.10.021
Rescuing dicer defects via inhibition of an anti-dicing nuclease
Abstract
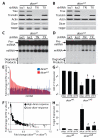
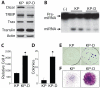
Genetic defects in the microRNA (miRNA) generating enzyme, dicer, are increasingly linked to disease. Loss of miRNA in dicer deficiency is thought to be due to loss of miRNA-generating activity. Here, we demonstrate a catabolic mechanism driving miRNA depletion in dicer deficiency. We developed a Dicer-antagonist assay revealing a pre-miRNA degrading enzyme that competes with pre-miRNA processing. We purified this pre-miRNA degrading activity using an unbiased chromatographic procedure and identified the ribonuclease complex Translin/Trax (TN/TX). In wild-type dicer backgrounds, pre-miRNA processing was dominant. However, in dicer-deficient contexts, TN/TX broadly suppressed miRNA. These findings indicate that miRNA depletion in dicer deficiency is due to the combined loss of miRNA-generating activity and catabolic function of TN/TX. Importantly, inhibition of TN/TX mitigated loss of both miRNA and tumor suppression with dicer haploinsufficiency. These studies reveal a potentially druggable target for restoring miRNA function in cancers and emerging dicer deficiencies.
Figures





References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nature genetics. 2003;35:215–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

