The bacterial nucleoid: nature, dynamics and sister segregation
- PMID: 25460806
- PMCID: PMC4359759
- DOI: 10.1016/j.mib.2014.10.001
The bacterial nucleoid: nature, dynamics and sister segregation
Erratum in
- Curr Opin Microbiol. 2015 Jun;25:146
Abstract
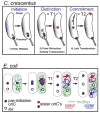
Recent studies reveal that the bacterial nucleoid has a defined, self-adherent shape and an underlying longitudinal organization and comprises a viscoelastic matrix. Within this shape, mobility is enhanced by ATP-dependent processes and individual loci can undergo ballistic off-equilibrium movements. In Escherichia coli, two global dynamic nucleoid behaviors emerge pointing to nucleoid-wide accumulation and relief of internal stress. Sister segregation begins with local splitting of individual loci, which is delayed at origin, terminus and specialized interstitial snap regions. Globally, as studied in several systems, segregation is a multi-step process in which internal nucleoid state plays critical roles that involve both compaction and expansion. The origin and terminus regions undergo specialized programs partially driven by complex ATP burning mechanisms such as a ParAB Brownian ratchet and a septum-associated FtsK motor. These recent findings reveal strong, direct parallels among events in different systems and between bacterial nucleoids and mammalian chromosomes with respect to physical properties, internal organization and dynamic behaviors.
Figures



References
-
- Jacob F, Brenner S. On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon. C R Hebd Seances Acad Sci. 1963;256:298–300. - PubMed
-
- Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol. 2012;86:1318–1333. Elucidation of nucleoid shape, its tendency for conservation over time and the significance of these features for physical models of bacterial genomes. - PMC - PubMed
-
- Fisher JK, Bourniquel A, Witz G, Weiner B, Prentiss M, Kleckner N. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153:882–895. Demonstration of efficient coalescence of chromosomal material into helical ellipsoid nucleoid shapes with underlying longitudinal duality. Modulation of nucleoid shape by cell shape, thus highlighting the importance of radial confinement. Identification of two global dynamic behaviors, both of which have correlates in mammalian cells: longitudinal density waves that oscillate on a time scale of minutes and global nucleoid extension and contraction cycles linked with release of programmed inter-sister links with characteristics of cyclic mechanical stress. Emphasis on the possible roles of non-specific and programmed tethers between chromosome segments for development of intra-nucleoid stress. - PMC - PubMed
-
- Umbarger MA, Toro E, Wright MA, Porreca GJ, Baù D, Hong S-H, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. Chromosome capture analysis defines, for the first time, organization of the bacterial nucleoid into a pair of relationally twisted longitudinal bundles. - PMC - PubMed
-
- Le TBK, Imakaev MV, Mirny LA, Laub MT. High-Resolution Mapping of the Spatial Organization of a Bacterial Chromosome. Science. 2013;342:731–734. First elucication of the detailed structure of a bacterial nucleoid. Left and right replichores of Caulobacter are each organized as a radial array of presumptive plectoneme loops enforced by nucleoid protein HU and longitudinally compacted by SMC. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

