In vivo continuous directed evolution
- PMID: 25461718
- PMCID: PMC4308500
- DOI: 10.1016/j.cbpa.2014.09.040
In vivo continuous directed evolution
Abstract
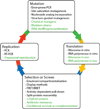
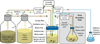
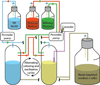
The development and application of methods for the laboratory evolution of biomolecules has rapidly progressed over the last few decades. Advancements in continuous microbe culturing and selection design have facilitated the development of new technologies that enable the continuous directed evolution of proteins and nucleic acids. These technologies have the potential to support the extremely rapid evolution of biomolecules with tailor-made functional properties. Continuous evolution methods must support all of the key steps of laboratory evolution - translation of genes into gene products, selection or screening, replication of genes encoding the most fit gene products, and mutation of surviving genes - in a self-sustaining manner that requires little or no researcher intervention. Continuous laboratory evolution has been historically used to study problems including antibiotic resistance, organismal adaptation, phylogenetic reconstruction, and host-pathogen interactions, with more recent applications focusing on the rapid generation of proteins and nucleic acids with useful, tailor-made properties. The advent of increasingly general methods for continuous directed evolution should enable researchers to address increasingly complex questions and to access biomolecules with more novel or even unprecedented properties.
Copyright © 2014. Published by Elsevier Ltd.
Figures

References
-
- Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. - PubMed
-
- Breaker RR, Banerji A, Joyce GF. Continuous in vitro evolution of bacteriophage RNA polymerase promoters. Biochemistry. 1994;33:11980–11986. - PubMed
-
- McGinness KE, Wright MC, Joyce GF. Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem Biol. 2002;9:585–596. - PubMed
-
- Kuhne H, Joyce GF. Continuous in vitro evolution of ribozymes that operate under conditions of extreme pH. J Mol Evol. 2003;57:292–298. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

