Proteome sequencing goes deep
- PMID: 25461719
- PMCID: PMC4308434
- DOI: 10.1016/j.cbpa.2014.10.017
Proteome sequencing goes deep
Abstract
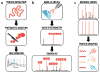
Advances in mass spectrometry (MS) have transformed the scope and impact of protein characterization efforts. Identifying hundreds of proteins from rather simple biological matrices, such as yeast, was a daunting task just a few decades ago. Now, expression of more than half of the estimated ∼20,000 human protein coding genes can be confirmed in record time and from minute sample quantities. Access to proteomic information at such unprecedented depths has been fueled by strides in every stage of the shotgun proteomics workflow-from sample processing to data analysis-and promises to revolutionize our understanding of the causes and consequences of proteome variation.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
References
-
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;2464:64–71. - PubMed
-
- Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet-laser desorption of nonvolatile compounds. Int J Mass Spectrom. 1987;78:53–68.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

