Computational schemes for the prediction and annotation of enhancers from epigenomic assays
- PMID: 25461775
- PMCID: PMC4778972
- DOI: 10.1016/j.ymeth.2014.10.008
Computational schemes for the prediction and annotation of enhancers from epigenomic assays
Abstract
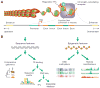
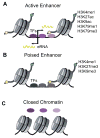
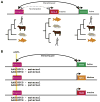
Identifying and annotating distal regulatory enhancers is critical to understand the mechanisms that control gene expression and cell-type-specific activities. Next-generation sequencing techniques have provided us an exciting toolkit of genome-wide assays that can be used to predict and annotate enhancers. However, each assay comes with its own specific set of analytical needs if enhancer prediction is to be optimal. Furthermore, integration of multiple genome-wide assays allows for different genomic features to be combined, and can improve predictive performance. Herein, we review the genome-wide assays and analysis schemes that are used to predict and annotate enhancers. In particular, we focus on three key computational topics: predicting enhancer locations, determining the cell-type-specific activity of enhancers, and linking enhancers to their target genes.
Keywords: Enhancer activity; Enhancer prediction; Enhancer–gene linking; Epigenetics; Epigenomics.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

References
-
- Smith E, Shilatifard A. Nat Struct Mol Biol. 2014;21:210–219. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

