Polymorphisms in DNA polymerase γ affect the mtDNA stability and the NRTI-induced mitochondrial toxicity in Saccharomyces cerevisiae
- PMID: 25462018
- PMCID: PMC4309887
- DOI: 10.1016/j.mito.2014.11.003
Polymorphisms in DNA polymerase γ affect the mtDNA stability and the NRTI-induced mitochondrial toxicity in Saccharomyces cerevisiae
Abstract
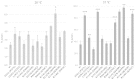
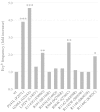
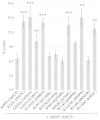
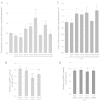
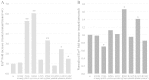
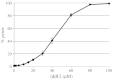
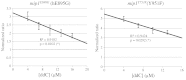
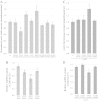
Several pathological mutations have been identified in human POLG gene, encoding for the catalytic subunit of Pol γ, the solely mitochondrial replicase in animals and fungi. However, little is known regarding non-pathological polymorphisms found in this gene. Here we studied, in the yeast model Saccharomyces cerevisiae, eight human polymorphisms. We found that most of them are not neutral but enhanced both mtDNA extended mutability and the accumulation of mtDNA point mutations, either alone or in combination with a pathological mutation. In addition, we found that the presence of some SNPs increased the stavudine and/or zalcitabine-induced mtDNA mutability and instability.
Keywords: MIP1; NRTI; POLG polymorphisms; Pharmacogenetics; Yeast model; mtDNA point and extended mutability.
Copyright © 2014. Published by Elsevier B.V.
Figures
References
-
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. vol. 2. Wiley; NY: 1994. Saccharomyces cerevisiae. (Current protocols in molecular biology).
-
- Azrak S., Ayyasamy V., Zirpoli G., Ambrosone C., Bandera E.V., Bovbjerg D.H., Jandorf L., Ciupak G., Davis W., Pawlish K.S., Liang P., Singh K. CAG repeat variants in the POLG1 gene encoding mtDNA polymerase-gamma and risk of breast cancer in African–American women. PLoS One. 2012;7:e29548. - PMC - PubMed
-
- Baruffini E., Lodi T. Construction and validation of a yeast model system for studying in vivo the susceptibility to nucleoside analogues of DNA polymerase gamma allelic variants. Mitochondrion. 2010;10:183–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

