Transgenic overexpression of mitofilin attenuates diabetes mellitus-associated cardiac and mitochondria dysfunction
- PMID: 25463274
- PMCID: PMC4302057
- DOI: 10.1016/j.yjmcc.2014.11.008
Transgenic overexpression of mitofilin attenuates diabetes mellitus-associated cardiac and mitochondria dysfunction
Abstract
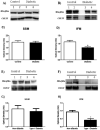
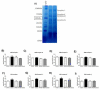
Mitofilin, also known as heart muscle protein, is an inner mitochondrial membrane structural protein that plays a central role in maintaining cristae morphology and structure. It is a critical component of the mitochondrial contact site and cristae organizing system (MICOS) complex which is important for mitochondrial architecture and cristae morphology. Our laboratory has previously reported alterations in mitochondrial morphology and proteomic make-up during type 1 diabetes mellitus, with mitofilin being significantly down-regulated in interfibrillar mitochondria (IFM). The goal of this study was to investigate whether overexpression of mitofilin can limit mitochondrial disruption associated with the diabetic heart through restoration of mitochondrial morphology and function. A transgenic mouse line overexpressing mitofilin was generated and mice injected intraperitoneally with streptozotocin using a multi low-dose approach. Five weeks following diabetes mellitus onset, cardiac contractile function was assessed. Restoration of ejection fraction and fractional shortening was observed in mitofilin diabetic mice as compared to wild-type controls (P<0.05 for both). Decrements observed in electron transport chain (ETC) complex I, III, IV and V activities, state 3 respiration, lipid peroxidation as well as mitochondria membrane potential in type 1 diabetic IFM were restored in mitofilin diabetic mice (P<0.05 for all). Qualitative analyses of electron micrographs revealed restoration of mitochondrial cristae structure in mitofilin diabetic mice as compared to wild-type controls. Furthermore, measurement of mitochondrial internal complexity using flow cytometry displayed significant reduction in internal complexity in diabetic IFM which was restored in mitofilin diabetic IFM (P<0.05). Taken together these results suggest that transgenic overexpression of mitofilin preserves mitochondrial structure, leading to restoration of mitochondrial function and attenuation of cardiac contractile dysfunction in the diabetic heart.
Keywords: Diabetes mellitus; Electron transport chain; Mitochondria; Mitofilin.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


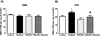



Comment in
-
Mitofilin: Key factor in diabetic cardiomyopathy?J Mol Cell Cardiol. 2015 Aug;85:292-3. doi: 10.1016/j.yjmcc.2014.11.028. Epub 2014 Dec 9. J Mol Cell Cardiol. 2015. PMID: 25500008 No abstract available.
References
-
- Mannella CA. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta. 2006 Feb;1762(2):140–7. - PubMed
-
- Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009 Jan;1793(1):5–19. - PubMed
-
- Gilkerson RW, Selker JM, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003 Jul 10;546(2-3):355–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR016440/RR/NCRR NIH HHS/United States
- T32 HL090610/HL/NHLBI NIH HHS/United States
- DP2 DK083095/DK/NIDDK NIH HHS/United States
- P30 RR032138/RR/NCRR NIH HHS/United States
- U54 GM104942/GM/NIGMS NIH HHS/United States
- DP2DK083095/DK/NIDDK NIH HHS/United States
- T32HL090610/HL/NHLBI NIH HHS/United States
- S10 RR026378/RR/NCRR NIH HHS/United States
- P30 RR031155/RR/NCRR NIH HHS/United States
- P30 GM103488/GM/NIGMS NIH HHS/United States
- U54GM104942/GM/NIGMS NIH HHS/United States
- P30RR031155/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

