Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China
- PMID: 25463600
- PMCID: PMC7112057
- DOI: 10.1016/j.virol.2014.10.017
Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China
Abstract
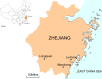
Although rodents are important reservoirs for RNA viruses, to date only one species of rodent coronavirus (CoV) has been identified. Herein, we describe a new CoV, denoted Lucheng Rn rat coronavirus (LRNV), and novel variants of two Betacoronavirus species termed Longquan Aa mouse coronavirus (LAMV) and Longquan Rl rat coronavirus (LRLV), that were identified in a survey of 1465 rodents sampled in China during 2011-2013. Phylogenetic analysis revealed that LAMV and LRLV fell into lineage A of the genus Betacoronavirus, which included CoVs discovered in humans and domestic and wild animals. In contrast, LRNV harbored by Rattus norvegicus formed a distinct lineage within the genus Alphacoronavirus in the 3CL(pro), RdRp, and Hel gene trees, but formed a more divergent lineage in the N and S gene trees, indicative of a recombinant origin. Additional recombination events were identified in LRLV. Together, these data suggest that rodents may carry additional unrecognized CoVs.
Keywords: Coronavirus; Evolution; Phylogeny; Recombination; Rodents.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Annan A., Baldwin H.J., Corman V.M., Klose S.M., Owusu M., Nkrumah E.E., Badu E.K., Anti P., Agbenyega O., Meyer B., Oppong S., Sarkodie Y.A., Kalko E.K., Lina P.H., Godlevska E.V., Reusken C., Seebens A., Gloza-Rausch F., Vallo P., Tschapka M., Drosten C., Drexler J.F. Human Betacoronavirus 2c EMC/2012-related viruses in bats. Ghana and Eur. Emerg. Infect. Dis. 2013;19:456–459. - PMC - PubMed
-
- Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N.J., Oinn T.M., Pagni M., Servant F., Sigrist C.J., Zdobnov E.M. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucl. Acids. Res. 2001;29:37–40. - PMC - PubMed
-
- Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370:2499–2505. - PubMed
-
- Beaudette F.R., Hudson C.B. Cultivation of the virus of infectious bronchitis. J. Am. Vet. Med. Assoc. 1937;90:51–58.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

