SPECHT - single-stage phosphopeptide enrichment and stable-isotope chemical tagging: quantitative phosphoproteomics of insulin action in muscle
- PMID: 25463755
- PMCID: PMC4289458
- DOI: 10.1016/j.jprot.2014.11.001
SPECHT - single-stage phosphopeptide enrichment and stable-isotope chemical tagging: quantitative phosphoproteomics of insulin action in muscle
Abstract
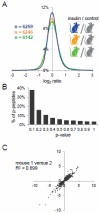
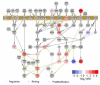
The study of cellular signaling remains a significant challenge for translational and clinical research. In particular, robust and accurate methods for quantitative phosphoproteomics in tissues and tumors represent significant hurdles for such efforts. In the present work, we design, implement and validate a method for single-stage phosphopeptide enrichment and stable isotope chemical tagging, or SPECHT, that enables the use of iTRAQ, TMT and/or reductive dimethyl-labeling strategies to be applied to phosphoproteomics experiments performed on primary tissue. We develop and validate our approach using reductive dimethyl-labeling and HeLa cells in culture, and find these results indistinguishable from data generated from more traditional SILAC-labeled HeLa cells mixed at the cell level. We apply the SPECHT approach to the quantitative analysis of insulin signaling in a murine myotube cell line and muscle tissue, identify known as well as new phosphorylation events, and validate these phosphorylation sites using phospho-specific antibodies. Taken together, our work validates chemical tagging post-single-stage phosphoenrichment as a general strategy for studying cellular signaling in primary tissues.
Biological significance: Through the use of a quantitatively reproducible, proteome-wide phosphopeptide enrichment strategy, we demonstrated the feasibility of post-phosphopeptide purification chemical labeling and tagging as an enabling approach for quantitative phosphoproteomics of primary tissues. Using reductive dimethyl labeling as a generalized chemical tagging strategy, we compared the performance of post-phosphopeptide purification chemical tagging to the well established community standard, SILAC, in insulin-stimulated tissue culture cells. We then extended our method to the analysis of low-dose insulin signaling in murine muscle tissue, and report on the analytical and biological significance of our results.
Keywords: Insulin signaling; Phosphoproteomics; Quantitative proteomics; Tissue proteomics.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
In-depth qualitative and quantitative profiling of tyrosine phosphorylation using a combination of phosphopeptide immunoaffinity purification and stable isotope dimethyl labeling.Mol Cell Proteomics. 2010 Jan;9(1):84-99. doi: 10.1074/mcp.M900291-MCP200. Epub 2009 Sep 21. Mol Cell Proteomics. 2010. PMID: 19770167 Free PMC article.
-
Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-Based Quantitative Proteomics and Phosphoproteomics in Fission Yeast.Cold Spring Harb Protoc. 2017 Jun 1;2017(6):pdb.prot091686. doi: 10.1101/pdb.prot091686. Cold Spring Harb Protoc. 2017. PMID: 28572185
-
Combining Metabolic ¹⁵N Labeling with Improved Tandem MOAC for Enhanced Probing of the Phosphoproteome.Methods Mol Biol. 2015;1306:81-96. doi: 10.1007/978-1-4939-2648-0_6. Methods Mol Biol. 2015. PMID: 25930695
-
Phosphoproteomics in cancer.Mol Oncol. 2010 Dec;4(6):482-95. doi: 10.1016/j.molonc.2010.09.004. Epub 2010 Sep 26. Mol Oncol. 2010. PMID: 20937571 Free PMC article. Review.
-
Advances in phosphopeptide enrichment techniques for phosphoproteomics.Amino Acids. 2012 Sep;43(3):1009-24. doi: 10.1007/s00726-012-1288-9. Epub 2012 Jul 22. Amino Acids. 2012. PMID: 22821267 Review.
Cited by
-
Distinct Hepatic PKA and CDK Signaling Pathways Control Activity-Independent Pyruvate Kinase Phosphorylation and Hepatic Glucose Production.Cell Rep. 2019 Dec 10;29(11):3394-3404.e9. doi: 10.1016/j.celrep.2019.11.009. Cell Rep. 2019. PMID: 31825824 Free PMC article.
-
Small changes in phospho-occupancy at the kinetochore-microtubule interface drive mitotic fidelity.J Cell Biol. 2022 Sep 5;221(9):e202107107. doi: 10.1083/jcb.202107107. Epub 2022 Jul 25. J Cell Biol. 2022. PMID: 35878017 Free PMC article.
-
Maintaining proteostasis under mechanical stress.EMBO Rep. 2021 Aug 4;22(8):e52507. doi: 10.15252/embr.202152507. Epub 2021 Jul 26. EMBO Rep. 2021. PMID: 34309183 Free PMC article. Review.
-
PKCε contributes to lipid-induced insulin resistance through cross talk with p70S6K and through previously unknown regulators of insulin signaling.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):E8996-E9005. doi: 10.1073/pnas.1804379115. Epub 2018 Sep 4. Proc Natl Acad Sci U S A. 2018. PMID: 30181290 Free PMC article.
-
Analysis of the Candida albicans Phosphoproteome.Eukaryot Cell. 2015 May;14(5):474-85. doi: 10.1128/EC.00011-15. Epub 2015 Mar 6. Eukaryot Cell. 2015. PMID: 25750214 Free PMC article.
References
-
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. - PubMed
-
- Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14(1):35–48. - PubMed
-
- Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134(2):353–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

