Influenza virus-host interactome screen as a platform for antiviral drug development
- PMID: 25464832
- PMCID: PMC4451456
- DOI: 10.1016/j.chom.2014.11.002
Influenza virus-host interactome screen as a platform for antiviral drug development
Abstract
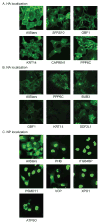
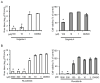
Host factors required for viral replication are ideal drug targets because they are less likely than viral proteins to mutate under drug-mediated selective pressure. Although genome-wide screens have identified host proteins involved in influenza virus replication, limited mechanistic understanding of how these factors affect influenza has hindered potential drug development. We conducted a systematic analysis to identify and validate host factors that associate with influenza virus proteins and affect viral replication. After identifying over 1,000 host factors that coimmunoprecipitate with specific viral proteins, we generated a network of virus-host protein interactions based on the stage of the viral life cycle affected upon host factor downregulation. Using compounds that inhibit these host factors, we validated several proteins, notably Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF1) and JAK1, as potential antiviral drug targets. Thus, virus-host interactome screens are powerful strategies to identify targetable host factors and guide antiviral drug development.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
-
- Boulo S, Akarsu H, Ruigrok RW, Baudin F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007;124:12–21. - PubMed
-
- Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. - PubMed
-
- Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

