An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes
- PMID: 25465409
- PMCID: PMC4311202
- DOI: 10.1105/tpc.114.131847
An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes
Abstract
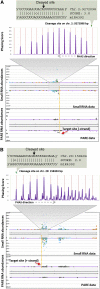
Small RNAs are ubiquitous, versatile repressors and include (1) microRNAs (miRNAs), processed from mRNA forming stem-loops; and (2) small interfering RNAs (siRNAs), the latter derived in plants by a process typically requiring an RNA-dependent RNA polymerase. We constructed and analyzed an expression atlas of soybean (Glycine max) small RNAs, identifying over 500 loci generating 21-nucleotide phased siRNAs (phasiRNAs; from PHAS loci), of which 483 overlapped annotated protein-coding genes. Via the integration of miRNAs with parallel analysis of RNA end (PARE) data, 20 miRNA triggers of 127 PHAS loci were detected. The primary class of PHAS loci (208 or 41% of the total) corresponded to NB-LRR genes; some of these small RNAs preferentially accumulate in nodules. Among the PHAS loci, novel representatives of TAS3 and noncanonical phasing patterns were also observed. A noncoding PHAS locus, triggered by miR4392, accumulated preferentially in anthers; the phasiRNAs are predicted to target transposable elements, with their peak abundance during soybean reproductive development. Thus, phasiRNAs show tremendous diversity in dicots. We identified novel miRNAs and assessed the veracity of soybean miRNAs registered in miRBase, substantially improving the soybean miRNA annotation, facilitating an improvement of miRBase annotations and identifying at high stringency novel miRNAs and their targets.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures





Comment in
-
A world beyond Arabidopsis: updates on small RNAs in plant development.Plant Cell. 2014 Dec;26(12):4564. doi: 10.1105/tpc.114.134635. Epub 2014 Dec 2. Plant Cell. 2014. PMID: 25465406 Free PMC article. No abstract available.
References
-
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. - PubMed
-
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

