miRNAs can be generally associated with human pathologies as exemplified for miR-144
- PMID: 25465851
- PMCID: PMC4268797
- DOI: 10.1186/s12916-014-0224-0
miRNAs can be generally associated with human pathologies as exemplified for miR-144
Abstract
Background: miRNA profiles are promising biomarker candidates for a manifold of human pathologies, opening new avenues for diagnosis and prognosis. Beyond studies that describe miRNAs frequently as markers for specific traits, we asked whether a general pattern for miRNAs across many diseases exists.
Methods: We evaluated genome-wide circulating profiles of 1,049 patients suffering from 19 different cancer and non-cancer diseases as well as unaffected controls. The results were validated on 319 individuals using qRT-PCR.
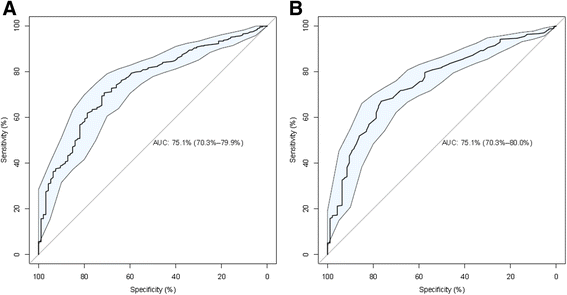
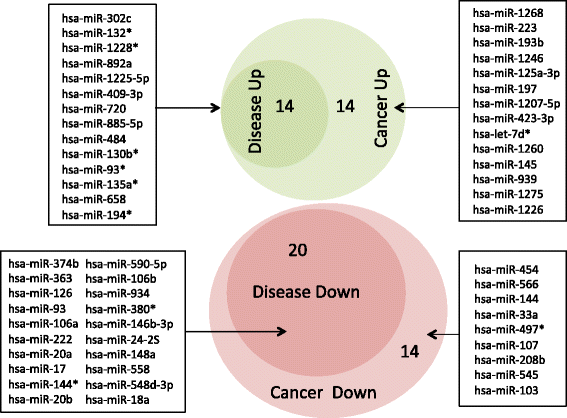
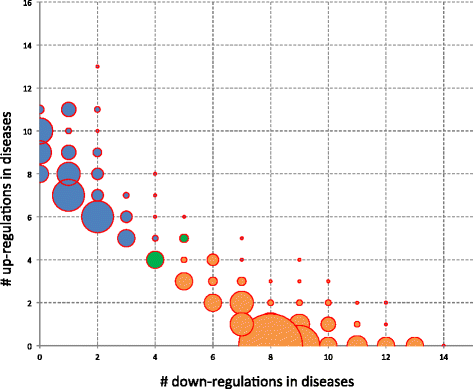
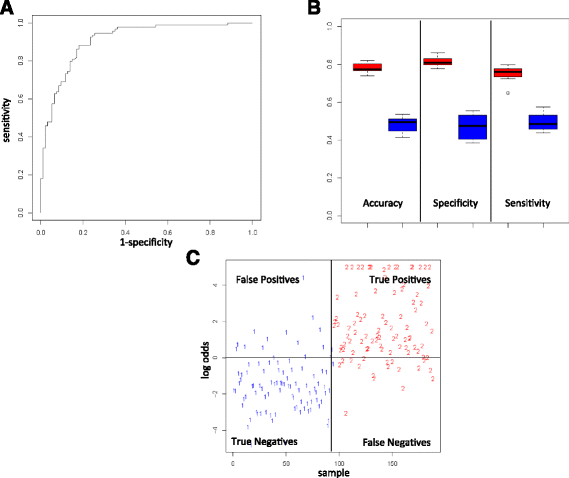
Results: We discovered 34 miRNAs with strong disease association. Among those, we found substantially decreased levels of hsa-miR-144* and hsa-miR-20b with AUC of 0.751 (95% CI: 0.703-0.799), respectively. We also discovered a set of miRNAs, including hsa-miR-155*, as rather stable markers, offering reasonable control miRNAs for future studies. The strong downregulation of hsa-miR-144* and the less variable pattern of hsa-miR-155* has been validated in a cohort of 319 samples in three different centers. Here, breast cancer as an additional disease phenotype not included in the screening phase has been included as the 20th trait.
Conclusions: Our study on 1,368 patients including 1,049 genome-wide miRNA profiles and 319 qRT-PCR validations further underscores the high potential of specific blood-borne miRNA patterns as molecular biomarkers. Importantly, we highlight 34 miRNAs that are generally dysregulated in human pathologies. Although these markers are not specific to certain diseases they may add to the diagnosis in combination with other markers, building a specific signature. Besides these dysregulated miRNAs, we propose a set of constant miRNAs that may be used as control markers.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

