Leading-process actomyosin coordinates organelle positioning and adhesion receptor dynamics in radially migrating cerebellar granule neurons
- PMID: 25467954
- PMCID: PMC4289176
- DOI: 10.1186/1749-8104-9-26
Leading-process actomyosin coordinates organelle positioning and adhesion receptor dynamics in radially migrating cerebellar granule neurons
Abstract
Background: During brain development, neurons migrate from germinal zones to their final positions to assemble neural circuits. A unique saltatory cadence involving cyclical organelle movement (e.g., centrosome motility) and leading-process actomyosin enrichment prior to nucleokinesis organizes neuronal migration. While functional evidence suggests that leading-process actomyosin is essential for centrosome motility, the role of the actin-enriched leading process in globally organizing organelle transport or traction forces remains unexplored.
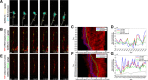
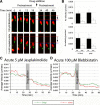
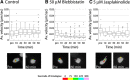
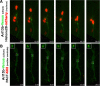
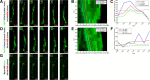
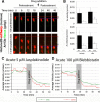
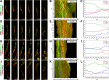
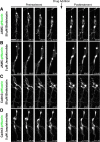
Results: We show that myosin ii motors and F-actin dynamics are required for Golgi apparatus positioning before nucleokinesis in cerebellar granule neurons (CGNs) migrating along glial fibers. Moreover, we show that primary cilia are motile organelles, localized to the leading-process F-actin-rich domain and immobilized by pharmacological inhibition of myosin ii and F-actin dynamics. Finally, leading process adhesion dynamics are dependent on myosin ii and F-actin.
Conclusions: We propose that actomyosin coordinates the overall polarity of migrating CGNs by controlling asymmetric organelle positioning and cell-cell contacts as these cells move along their glial guides.
Figures

References
-
- LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Dev Neurosci. 2006;29:407–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

