Self-organizing actomyosin patterns on the cell cortex at epithelial cell-cell junctions
- PMID: 25468344
- PMCID: PMC4255218
- DOI: 10.1016/j.bpj.2014.10.045
Self-organizing actomyosin patterns on the cell cortex at epithelial cell-cell junctions
Abstract
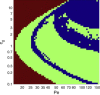
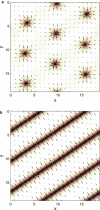
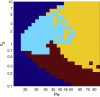
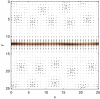
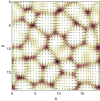
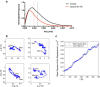
The behavior of actomyosin critically determines morphologically distinct patterns of contractility found at the interface between adherent cells. One such pattern is found at the apical region (zonula adherens) of cell-cell junctions in epithelia, where clusters of the adhesion molecule E-cadherin concentrate in a static pattern. Meanwhile, E-cadherin clusters throughout lateral cell-cell contacts display dynamic movements in the plane of the junctions. To gain insight into the principles that determine the nature and organization of these dynamic structures, we analyze this behavior by modeling the 2D actomyosin cell cortex as an active fluid medium. The numerical simulations show that the stability of the actin filaments influences the spatial structure and dynamics of the system. We find that in addition to static Turing-type patterns, persistent dynamic behavior occurs in a wide range of parameters. In the 2D model, mechanical stress-dependent actin breakdown is shown to produce a continuously changing network of actin bridges, whereas with a constant breakdown rate, more isolated clusters of actomyosin tend to form. The model qualitatively reproduces the dynamic and stable patterns experimentally observed at the junctions between epithelial cells.
Copyright © 2014 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Gierer A., Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. - PubMed
-
- Kondo S., Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. - PubMed
-
- Sick S., Reinker S., Schlake T. WNT and DKK determine hair follicle spacing through a reaction-diffusion mechanism. Science. 2006;314:1447–1450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

