Expression of pluripotency factors in echinoderm regeneration
- PMID: 25468557
- PMCID: PMC4323854
- DOI: 10.1007/s00441-014-2040-4
Expression of pluripotency factors in echinoderm regeneration
Abstract
Cell dedifferentiation is an integral component of post-traumatic regeneration in echinoderms. As dedifferentiated cells become multipotent, we asked if this spontaneous broadening of developmental potential is associated with the action of the same pluripotency factors (known as Yamanaka factors) that were used to induce pluripotency in specialized mammalian cells. In this study, we investigate the expression of orthologs of the four Yamanaka factors in regeneration of two different organs, the radial nerve cord and the digestive tube, in the sea cucumber Holothuria glaberrima. All four pluripotency factors are expressed in uninjured animals, although their expression domains do not always overlap. In regeneration, the expression levels of the four genes were not regulated in a coordinated way, but instead showed different dynamics for individual genes and also were different between the radial nerve and the gut. SoxB1, the ortholog of the mammalian Sox2, was drastically downregulated in the regenerating intestine, suggesting that this factor is not required for dedifferentiation/regeneration in this organ. On the other hand, during the early post-injury stage, Myc, the sea cucumber ortholog of c-Myc, was significantly upregulated in both the intestine and the radial nerve cord and is therefore hypothesized to play a central role in dedifferentiation/regeneration of various tissue types.
Figures



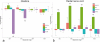




References
-
- Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, Neufeld K, Clevers H, Brunton V, Winton DJ, Wang X, Sears RC, Clarke AR, Frame MC, Sansom OJ. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell. 2010;19(2):259–269. DOI 10.1016/j.devcel.2010.07.015, URL http://dx.doi.org/10.1016/j.devcel.2010.07.015. - DOI - PMC - PubMed
-
- Bhavsar RB, Tsonis PA. Exogenous Oct-4 inhibits lens transdifferentiation in the newt Notophthalmus viridescens. PLoS One. 2014;9(7):e102–510. DOI 0, URL http://dx.doi.org/0. - PMC - PubMed
-
- Brockes JP, Gates PB. Mechanisms underlying vertebrate limb regeneration: lessons from the salamander. Biochem Soc Trans. 2014;42(3):625–630. DOI 10.1042/BST20140002, URL http://dx.doi.org/10.1042/BST20140002. - DOI - PubMed
-
- Candelaria AG, Murray G, File SK, García-Arrarás JE. Contribution of mesenterial muscle dedifferentiation to intestine regeneration in the sea cucumber Holothuria glaberrima. Cell Tissue Res. 2006;325(1):55–65. DOI 10.1007/s00441-006-0170-z, URL http://dx.doi.org/10.1007/s00441-006-0170-z. - DOI - PubMed
-
- Christen B, Robles V, Raya M, Paramonov I, Izpisuá Belmonte JC. Regeneration and reprogramming compared. BMC Biol. 2010;8:5. DOI 10.1186/1741-7007-8-5, URL http://dx.doi.org/10.1186/1741-7007-8-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

