Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and MiRNA profiles
- PMID: 25469788
- PMCID: PMC4254602
- DOI: 10.1371/journal.pone.0113298
Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and MiRNA profiles
Abstract
Background: Whole blood expression profiling is frequently performed using PAXgene (Qiagen) or Tempus (Life Technologies) tubes. Here, we compare 6 novel generation RNA isolation protocols with respect to RNA quantity, quality and recovery of mRNA and miRNA.
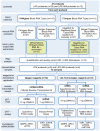
Methods: 3 PAXgene and 3 Tempus Tubes were collected from participants of the LIFE study with (n = 12) and without (n = 35) acute myocardial infarction (AMI). RNA was extracted with 4 manual protocols from Qiagen (PAXgene Blood miRNA Kit), Life Technologies (MagMAX for Stabilized Blood Tubes RNA Isolation Kit), and Norgen Biotek (Norgen Preserved Blood RNA Purification Kit I and Kit II), and 2 (semi-)automated protocols on the QIAsymphony (Qiagen) and MagMAX Express-96 Magnetic Particle Processor (Life Technologies). RNA quantity and quality was determined. For biological validation, RNA from 12 representative probands, extracted with all 6 kits (n = 72), was reverse transcribed and mRNAs (matrix metalloproteinase 9, arginase 1) and miRNAs (miR133a, miR1), shown to be altered by AMI, were analyzed.
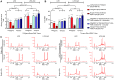
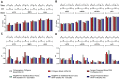
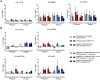
Results: RNA yields were highest using the Norgen Kit I with Tempus Tubes and lowest using the Norgen Kit II with PAXgene. The disease status was the second major determinant of RNA yields (LIFE-AMI 11.2 vs. LIFE 6.7 µg, p<0.001) followed by the choice of blood collection tube. (Semi-)automation reduced overall RNA extraction time but did not generally reduce hands-on-time. RNA yields and quality were comparable between manual and automated extraction protocols. mRNA expression was not affected by collection tubes and RNA extraction kits but by RT/qPCR reagents with exception of the Norgen Kit II, which led to mRNA depletion. For miRNAs, expression differences related to collection tubes (miR30b), RNA isolation (Norgen Kit II), and RT/qRT reagents (miR133a) were observed.
Conclusion: We demonstrate that novel generation RNA isolation kits significantly differed with respect to RNA recovery and affected miRNA but not mRNA expression profiles.
Conflict of interest statement
Figures
References
-
- Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, et al. (2002) Stabilization of mRNA expression in whole blood samples. Clin Chem 48:1883–1890. - PubMed
-
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Moser K, et al. (2004) Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes and Immunity 5:347–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

