Structure of a mammalian ryanodine receptor
- PMID: 25470061
- PMCID: PMC4300236
- DOI: 10.1038/nature13950
Structure of a mammalian ryanodine receptor
Abstract
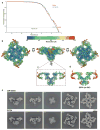
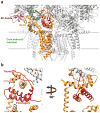
Ryanodine receptors (RyRs) mediate the rapid release of calcium (Ca(2+)) from intracellular stores into the cytosol, which is essential for numerous cellular functions including excitation-contraction coupling in muscle. Lack of sufficient structural detail has impeded understanding of RyR gating and regulation. Here we report the closed-state structure of the 2.3-megadalton complex of the rabbit skeletal muscle type 1 RyR (RyR1), solved by single-particle electron cryomicroscopy at an overall resolution of 4.8 Å. We fitted a polyalanine-level model to all 3,757 ordered residues in each protomer, defining the transmembrane pore in unprecedented detail and placing all cytosolic domains as tertiary folds. The cytosolic assembly is built on an extended α-solenoid scaffold connecting key regulatory domains to the pore. The RyR1 pore architecture places it in the six-transmembrane ion channel superfamily. A unique domain inserted between the second and third transmembrane helices interacts intimately with paired EF-hands originating from the α-solenoid scaffold, suggesting a mechanism for channel gating by Ca(2+).
Conflict of interest statement
The authors declare no competing financial interests.
Figures








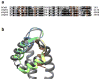
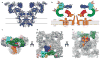


References
-
- Zalk R, Lehnart SE, Marks AR. Modulation of the ryanodine receptor and intracellular calcium. Annual review of biochemistry. 2007;76:367–385. - PubMed
-
- Marx SO, et al. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. - PubMed
-
- Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. Journal of molecular biology. 2004;342:145–153. - PubMed
-
- Meissner G, Lu X. Dihydropyridine receptor-ryanodine receptor interactions in skeletal muscle excitation-contraction coupling. Bioscience reports. 1995;15:399–408. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

