Structure of the F-actin-tropomyosin complex
- PMID: 25470062
- PMCID: PMC4477711
- DOI: 10.1038/nature14033
Structure of the F-actin-tropomyosin complex
Abstract
Filamentous actin (F-actin) is the major protein of muscle thin filaments, and actin microfilaments are the main component of the eukaryotic cytoskeleton. Mutations in different actin isoforms lead to early-onset autosomal dominant non-syndromic hearing loss, familial thoracic aortic aneurysms and dissections, and multiple variations of myopathies. In striated muscle fibres, the binding of myosin motors to actin filaments is mainly regulated by tropomyosin and troponin. Tropomyosin also binds to F-actin in smooth muscle and in non-muscle cells and stabilizes and regulates the filaments there in the absence of troponin. Although crystal structures for monomeric actin (G-actin) are available, a high-resolution structure of F-actin is still missing, hampering our understanding of how disease-causing mutations affect the function of thin muscle filaments and microfilaments. Here we report the three-dimensional structure of F-actin at a resolution of 3.7 Å in complex with tropomyosin at a resolution of 6.5 Å, determined by electron cryomicroscopy. The structure reveals that the D-loop is ordered and acts as a central region for hydrophobic and electrostatic interactions that stabilize the F-actin filament. We clearly identify map density corresponding to ADP and Mg(2+) and explain the possible effect of prominent disease-causing mutants. A comparison of F-actin with G-actin reveals the conformational changes during filament formation and identifies the D-loop as their key mediator. We also confirm that negatively charged tropomyosin interacts with a positively charged groove on F-actin. Comparison of the position of tropomyosin in F-actin-tropomyosin with its position in our previously determined F-actin-tropomyosin-myosin structure reveals a myosin-induced transition of tropomyosin. Our results allow us to understand the role of individual mutations in the genesis of actin- and tropomyosin-related diseases and will serve as a strong foundation for the targeted development of drugs.
Figures

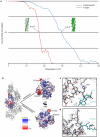
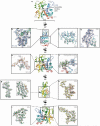








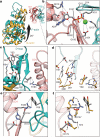
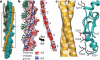
References
-
- Milewicz DM, et al. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu. Rev. Genomics Hum. Genet. 2008;9:283–302. - PubMed
-
- Sparrow JC, et al. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 2003;13:519–531. - PubMed
-
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. - PubMed
-
- Maytum R, Lehrer SS, Geeves MA. Cooperativity and switching within the three-state model of muscle regulation. Biochemistry. 1999;38:1102–1110. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

