Deconstructing transcriptional heterogeneity in pluripotent stem cells
- PMID: 25471879
- PMCID: PMC4256722
- DOI: 10.1038/nature13920
Deconstructing transcriptional heterogeneity in pluripotent stem cells
Abstract
Pluripotent stem cells (PSCs) are capable of dynamic interconversion between distinct substates; however, the regulatory circuits specifying these states and enabling transitions between them are not well understood. Here we set out to characterize transcriptional heterogeneity in mouse PSCs by single-cell expression profiling under different chemical and genetic perturbations. Signalling factors and developmental regulators show highly variable expression, with expression states for some variable genes heritable through multiple cell divisions. Expression variability and population heterogeneity can be influenced by perturbation of signalling pathways and chromatin regulators. Notably, either removal of mature microRNAs or pharmacological blockage of signalling pathways drives PSCs into a low-noise ground state characterized by a reconfigured pluripotency network, enhanced self-renewal and a distinct chromatin state, an effect mediated by opposing microRNA families acting on the Myc/Lin28/let-7 axis. These data provide insight into the nature of transcriptional heterogeneity in PSCs.
Figures
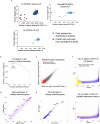


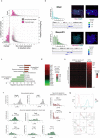


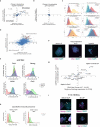
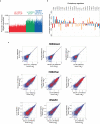
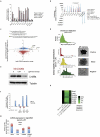
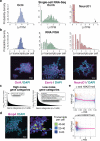
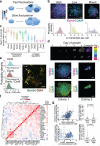
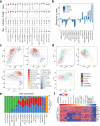

Comment in
-
GENE EXPRESSION. Single-cell variability guided by microRNAs.Science. 2016 Jun 17;352(6292):1390-1. doi: 10.1126/science.aag1097. Science. 2016. PMID: 27313022 No abstract available.
References
-
- Loh Y-H, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–440. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- T32 HL066987/HL/NHLBI NIH HHS/United States
- P50 HG005550/HG/NHGRI NIH HHS/United States
- F32 HD075541/HD/NICHD NIH HHS/United States
- DP1 CA174427/CA/NCI NIH HHS/United States
- K01 DK096013/DK/NIDDK NIH HHS/United States
- P50 HG006193/HG/NHGRI NIH HHS/United States
- T32 HL007623/HL/NHLBI NIH HHS/United States
- DP1 OD003958/OD/NIH HHS/United States
- R01 GM107536/GM/NIGMS NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R24 DK092760/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

