Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication
- PMID: 25473060
- PMCID: PMC4338866
- DOI: 10.1128/JVI.03073-14
Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication
Abstract
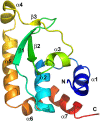
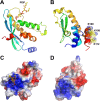
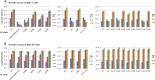
Phosphoinositides and phosphoinositide binding proteins play a critical role in membrane and protein trafficking in eukaryotes. Their critical role in replication of cytoplasmic viruses has just begun to be understood. Poxviruses, a family of large cytoplasmic DNA viruses, rely on the intracellular membranes to develop their envelope, and poxvirus morphogenesis requires enzymes from the cellular phosphoinositide metabolic pathway. However, the role of phosphoinositides in poxvirus replication remains unclear, and no poxvirus proteins show any homology to eukaryotic phosphoinositide binding domains. Recently, a group of poxvirus proteins, termed viral membrane assembly proteins (VMAPs), were identified as essential for poxvirus membrane biogenesis. A key component of VMAPs is the H7 protein. Here we report the crystal structure of the H7 protein from vaccinia virus. The H7 structure displays a novel fold comprised of seven α-helices and a highly curved three-stranded antiparallel β-sheet. We identified a phosphoinositide binding site in H7, comprised of basic residues on a surface patch and the flexible C-terminal tail. These residues were found to be essential for viral replication and for binding of H7 to phosphatidylinositol-3-phosphate (PI3P) and phosphatidylinositol-4-phosphate (PI4P). Our studies suggest that phosphoinositide binding by H7 plays an essential role in poxvirus membrane biogenesis.
Importance: Poxvirus viral membrane assembly proteins (VMAPs) were recently shown to be essential for poxvirus membrane biogenesis. One of the key components of VMAPs is the H7 protein. However, no known structural motifs could be identified from its sequence, and there are no homologs of H7 outside the poxvirus family to suggest a biochemical function. We have determined the crystal structure of the vaccinia virus (VACV) H7 protein. The structure displays a novel fold with a distinct and positively charged surface. Our data demonstrate that H7 binds phosphatidylinositol-3-phosphate and phosphatidylinositol-4-phosphate and that the basic surface patch is indeed required for phosphoinositide binding. In addition, mutation of positively charged residues required for lipid binding disrupted VACV replication. Phosphoinositides and phosphoinositide binding proteins play critical roles in membrane and protein trafficking in eukaryotes. Our study demonstrates that VACV H7 displays a novel fold for phosphoinositide binding, which is essential for poxvirus replication.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



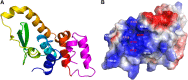
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

