Chromatin accessibility: a window into the genome
- PMID: 25473421
- PMCID: PMC4253006
- DOI: 10.1186/1756-8935-7-33
Chromatin accessibility: a window into the genome
Abstract
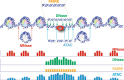
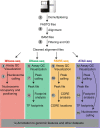
Transcriptional activation throughout the eukaryotic lineage has been tightly linked with disruption of nucleosome organization at promoters, enhancers, silencers, insulators and locus control regions due to transcription factor binding. Regulatory DNA thus coincides with open or accessible genomic sites of remodeled chromatin. Current chromatin accessibility assays are used to separate the genome by enzymatic or chemical means and isolate either the accessible or protected locations. The isolated DNA is then quantified using a next-generation sequencing platform. Wide application of these assays has recently focused on the identification of the instrumental epigenetic changes responsible for differential gene expression, cell proliferation, functional diversification and disease development. Here we discuss the limitations and advantages of current genome-wide chromatin accessibility assays with especial attention on experimental precautions and sequence data analysis. We conclude with our perspective on future improvements necessary for moving the field of chromatin profiling forward.
Keywords: ATAC; Chromatin; DNase; Epigenome; FAIRE; Histone; Library; MNase; Nucleosome; Sequencing.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

