Enhancer variants: evaluating functions in common disease
- PMID: 25473424
- PMCID: PMC4254432
- DOI: 10.1186/s13073-014-0085-3
Enhancer variants: evaluating functions in common disease
Abstract
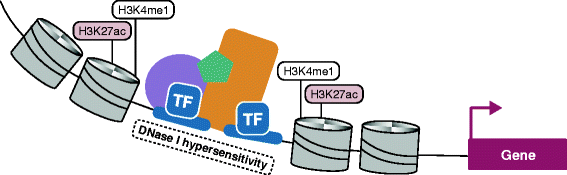
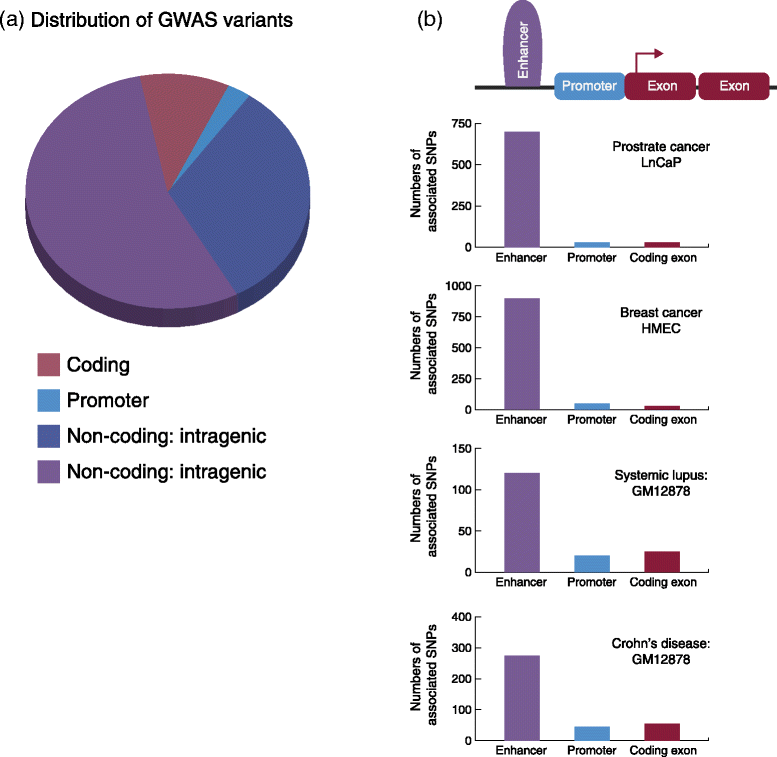
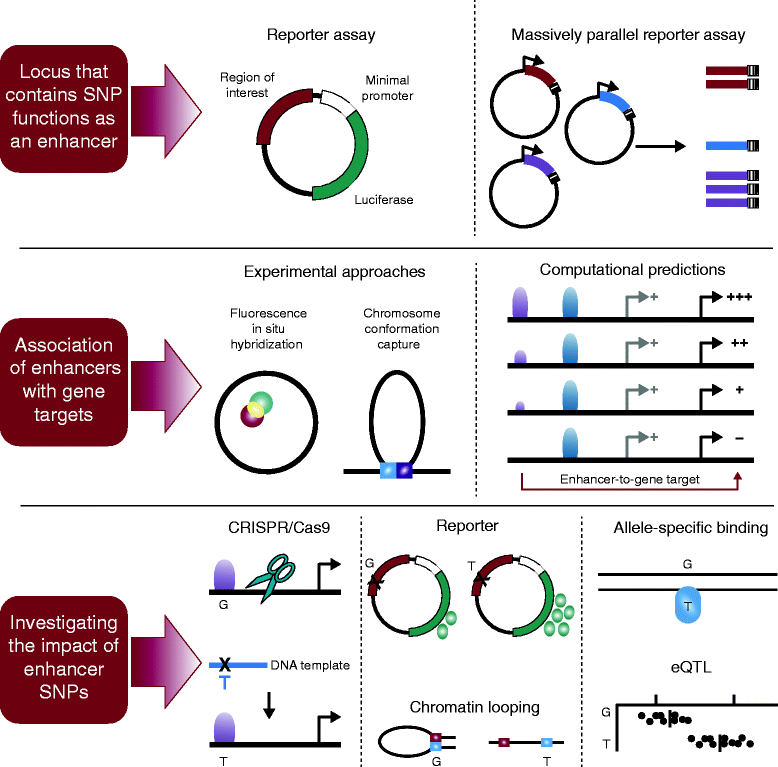
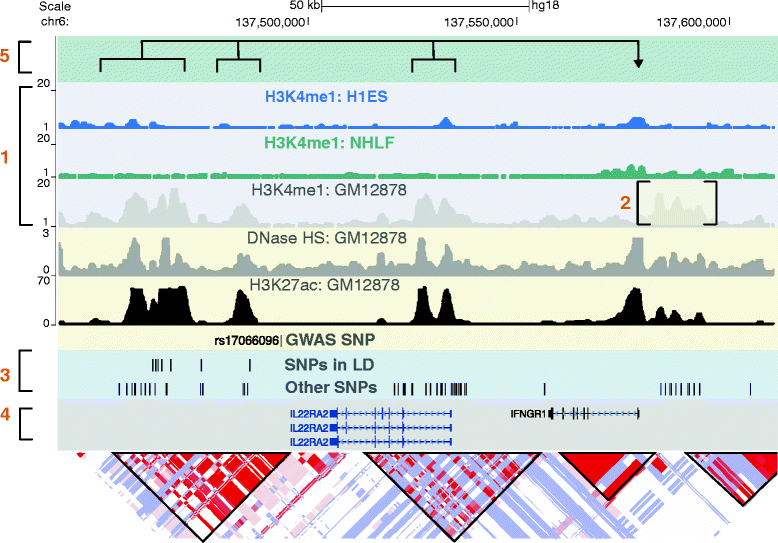
Gene enhancer elements are noncoding segments of DNA that play a central role in regulating transcriptional programs that control development, cell identity, and evolutionary processes. Recent studies have shown that noncoding single nucleotide polymorphisms (SNPs) that have been associated with risk for numerous common diseases through genome-wide association studies frequently lie in cell-type-specific enhancer elements. These enhancer variants probably influence transcriptional output, thereby offering a mechanistic basis to explain their association with risk for many common diseases. This review focuses on the identification and interpretation of disease-susceptibility variants that influence enhancer function. We discuss strategies for prioritizing the study of functional enhancer SNPs over those likely to be benign, review experimental and computational approaches to identifying the gene targets of enhancer variants, and highlight efforts to quantify the impact of enhancer variants on target transcript levels and cellular phenotypes. These studies are beginning to provide insights into the mechanistic basis of many common diseases, as well as into how we might translate this knowledge for improved disease diagnosis, prevention and treatments. Finally, we highlight five major challenges often associated with interpreting enhancer variants, and discuss recent technical advances that may help to surmount these challenges.
Figures
References
-
- A Catalog of Published Genome-Wide Association Studies. [http://www.genome.gov/gwasstudies]
-
- Gusev A, Hong Lee S, Neale BM, Trynka G, Vilhjalmsson BJ, Finucane H, Xu H, Zang C, Ripka S, Stahl E, Schizophrenia Working Group of the PGC, SWE-SCZ Consortium. Kahler AK, Hultman CM, Purcell SM, McCarroll SA, Daly M, Pasaniuc B, Sullivan PF, Wray NR, Raychaudhuri S, Price AL. Regulatory variants explain much more heritability than coding variants across 11 common diseases. bioRxiv. 2014 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

