A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis
- PMID: 25474254
- PMCID: PMC4256081
- DOI: 10.1371/journal.pcbi.1004016
A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis
Erratum in
-
Correction: A Systematic Computational Analysis of Biosynthetic Gene Cluster Evolution: Lessons for Engineering Biosynthesis.PLoS Comput Biol. 2016 Mar 8;12(3):e1004767. doi: 10.1371/journal.pcbi.1004767. eCollection 2016 Mar. PLoS Comput Biol. 2016. PMID: 26953826 Free PMC article. No abstract available.
Abstract
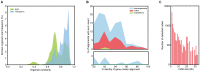
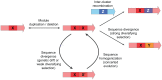
Bacterial secondary metabolites are widely used as antibiotics, anticancer drugs, insecticides and food additives. Attempts to engineer their biosynthetic gene clusters (BGCs) to produce unnatural metabolites with improved properties are often frustrated by the unpredictability and complexity of the enzymes that synthesize these molecules, suggesting that genetic changes within BGCs are limited by specific constraints. Here, by performing a systematic computational analysis of BGC evolution, we derive evidence for three findings that shed light on the ways in which, despite these constraints, nature successfully invents new molecules: 1) BGCs for complex molecules often evolve through the successive merger of smaller sub-clusters, which function as independent evolutionary entities. 2) An important subset of polyketide synthases and nonribosomal peptide synthetases evolve by concerted evolution, which generates sets of sequence-homogenized domains that may hold promise for engineering efforts since they exhibit a high degree of functional interoperability, 3) Individual BGC families evolve in distinct ways, suggesting that design strategies should take into account family-specific functional constraints. These findings suggest novel strategies for using synthetic biology to rationally engineer biosynthetic pathways.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: MAF is on the scientific advisory board of Warp Drive Bio.
Figures


References
-
- Osbourn A (2010) Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet 26: 449–457. - PubMed
-
- Sherman DH (2005) The Lego-ization of polyketide biosynthesis. Nat Biotechnol 23: 1083–1084. - PubMed
-
- Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, et al. (2005) Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol 23: 1171–1176. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

