Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis
- PMID: 25474325
- PMCID: PMC4256289
- DOI: 10.1371/journal.pntd.0003348
Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis
Abstract
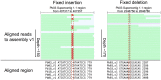
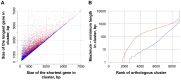
Paracoccidiodomycosis (PCM) is a clinically important fungal disease that can acquire serious systemic forms and is caused by the thermodimorphic fungal Paracoccidioides spp. PCM is a tropical disease that is endemic in Latin America, where up to ten million people are infected; 80% of reported cases occur in Brazil, followed by Colombia and Venezuela. To enable genomic studies and to better characterize the pathogenesis of this dimorphic fungus, two reference strains of P. brasiliensis (Pb03, Pb18) and one strain of P. lutzii (Pb01) were sequenced [1]. While the initial draft assemblies were accurate in large scale structure and had high overall base quality, the sequences had frequent small scale defects such as poor quality stretches, unknown bases (N's), and artifactual deletions or nucleotide duplications, all of which caused larger scale errors in predicted gene structures. Since assembly consensus errors can now be addressed using next generation sequencing (NGS) in combination with recent methods allowing systematic assembly improvement, we re-sequenced the three reference strains of Paracoccidioides spp. using Illumina technology. We utilized the high sequencing depth to re-evaluate and improve the original assemblies generated from Sanger sequence reads, and obtained more complete and accurate reference assemblies. The new assemblies led to improved transcript predictions for the vast majority of genes of these reference strains, and often substantially corrected gene structures. These include several genes that are central to virulence or expressed during the pathogenic yeast stage in Paracoccidioides and other fungi, such as HSP90, RYP1-3, BAD1, catalase B, alpha-1,3-glucan synthase and the beta glucan synthase target gene FKS1. The improvement and validation of these reference sequences will now allow more accurate genome-based analyses. To our knowledge, this is one of the first reports of a fully automated and quality-assessed upgrade of a genome assembly and annotation for a non-model fungus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

