The role of the gut microbiota in the pathogenesis of antiphospholipid syndrome
- PMID: 25475595
- PMCID: PMC4394866
- DOI: 10.1007/s11926-014-0472-1
The role of the gut microbiota in the pathogenesis of antiphospholipid syndrome
Abstract
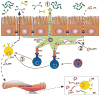
Infectious triggers are associated with the induction of transient antiphospholipid antibodies. One therefore wonders if microbes that permanently colonize us play a role in the pathogenesis of antiphospholipid syndrome (APS). The microbiota represents the collection of all microorganisms colonizing humans and is necessary for normal host physiology. The microbiota, however, is a constant stress on the immune system, which is tasked with recognizing and eliminating pathogenic microbes while tolerating commensal populations. A growing body of literature supports a critical role for the commensal-immune axis in the development of autoimmunity against colonized barriers (e.g., gut or skin) and sterile organs (e.g., pancreas or joints). Whether these interactions affect the development and sustainment of autoreactive CD4(+) T cells and pathogenic autoantibodies in APS is unknown. This review provides an overview of the current understanding of the commensal-immune axis in autoimmunity with a focus on the potential relevance to APS. Additionally, we discuss emerging findings supporting the involvement of the gut microbiota in a spontaneous model of APS, the (NZW × BXSB)F1 hybrid, and formalize hypotheses to explain how interactions between the immune system and the microbiota may influence human APS etiopathogenesis.
Conflict of interest statement
Figures
References
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

