Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis
- PMID: 25475669
- PMCID: PMC4281000
- DOI: 10.1104/pp.114.247767
Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis
Abstract
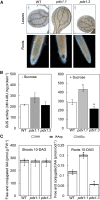
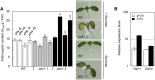
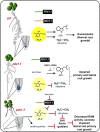
Vitamin B(6) (pyridoxal 5'-phosphate) is an essential cofactor of many metabolic enzymes. Plants biosynthesize the vitamin de novo employing two enzymes, pyridoxine synthase1 (PDX1) and PDX2. In Arabidopsis (Arabidopsis thaliana), there are two catalytically active paralogs of PDX1 (PDX1.1 and PDX1.3) producing the vitamin at comparable rates. Since single mutants are viable but the pdx1.1 pdx1.3 double mutant is lethal, the corresponding enzymes seem redundant. However, the single mutants exhibit substantial phenotypic differences, particularly at the level of root development, with pdx1.3 being more impaired than pdx1.1. Here, we investigate the differential regulation of PDX1.1 and PDX1.3 by identifying factors involved in their disparate phenotypes. Swapped-promoter experiments clarify the presence of distinct regulatory elements in the upstream regions of both genes. Exogenous sucrose (Suc) triggers impaired ethylene production in both mutants but is more severe in pdx1.3 than in pdx1.1. Interestingly, Suc specifically represses PDX1.1 expression, accounting for the stronger vitamin B6 deficit in pdx1.3 compared with pdx1.1. Surprisingly, Suc enhances auxin levels in pdx1.1, whereas the levels are diminished in pdx1.3. In the case of pdx1.3, the previously reported reduced meristem activity combined with the impaired ethylene and auxin levels manifest the specific root developmental defects. Moreover, it is the deficit in ethylene production and/or signaling that triggers this outcome. On the other hand, we hypothesize that it is the increased auxin content of pdx1.1 that is responsible for the root developmental defects observed therein. We conclude that PDX1.1 and PDX1.3 play partially nonredundant roles and are differentially regulated as manifested in disparate root growth impairment morphologies.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis.Plant J. 2006 Dec;48(6):933-46. doi: 10.1111/j.1365-313X.2006.02928.x. Plant J. 2006. PMID: 17227548
-
Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses.Plant J. 2005 Nov;44(3):396-408. doi: 10.1111/j.1365-313X.2005.02538.x. Plant J. 2005. PMID: 16236150
-
Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 protein family in metabolism, development, and vitamin B6 biosynthesis.Plant Cell. 2006 Jul;18(7):1722-35. doi: 10.1105/tpc.105.036269. Epub 2006 Jun 9. Plant Cell. 2006. PMID: 16766694 Free PMC article.
-
Auxin-mediated lateral root formation in higher plants.Int Rev Cytol. 2007;256:111-37. doi: 10.1016/S0074-7696(07)56004-3. Int Rev Cytol. 2007. PMID: 17241906 Review.
-
Two independent routes of de novo vitamin B6 biosynthesis: not that different after all.Biochem J. 2007 Oct 1;407(1):1-13. doi: 10.1042/BJ20070765. Biochem J. 2007. PMID: 17822383 Review.
Cited by
-
Plant Growth Regulation in Cell and Tissue Culture In Vitro.Plants (Basel). 2024 Jan 22;13(2):327. doi: 10.3390/plants13020327. Plants (Basel). 2024. PMID: 38276784 Free PMC article. Review.
-
Small kernel2 Encodes a Glutaminase in Vitamin B6 Biosynthesis Essential for Maize Seed Development.Plant Physiol. 2017 Jun;174(2):1127-1138. doi: 10.1104/pp.16.01295. Epub 2017 Apr 13. Plant Physiol. 2017. PMID: 28408540 Free PMC article.
-
Temporal Shifts in Hormone Signaling Networks Orchestrate Soybean Floral Development Under Field Conditions: An RNA-Seq Study.Int J Mol Sci. 2025 Jul 4;26(13):6455. doi: 10.3390/ijms26136455. Int J Mol Sci. 2025. PMID: 40650230 Free PMC article.
-
Transcriptome Analysis Revealed the Regulatory Mechanism of DIMBOA Affecting Early Somatic Embryogenesis in Dimocarpus longan Lour.Plants (Basel). 2025 Feb 3;14(3):442. doi: 10.3390/plants14030442. Plants (Basel). 2025. PMID: 39943004 Free PMC article.
-
Genome-wide identification of PDX and expression analysis under waterlogging stress exhibit stronger waterlogging tolerance in transgenic Brassica napus plants overexpressing the BnaPDX1.3 gene compared to wild-type plants.Front Plant Sci. 2025 Feb 12;16:1533219. doi: 10.3389/fpls.2025.1533219. eCollection 2025. Front Plant Sci. 2025. PMID: 40012725 Free PMC article.
References
-
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 - PubMed
-
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
-
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

