Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse
- PMID: 25476147
- PMCID: PMC4295324
- DOI: 10.1186/s13059-014-0545-5
Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse
Abstract
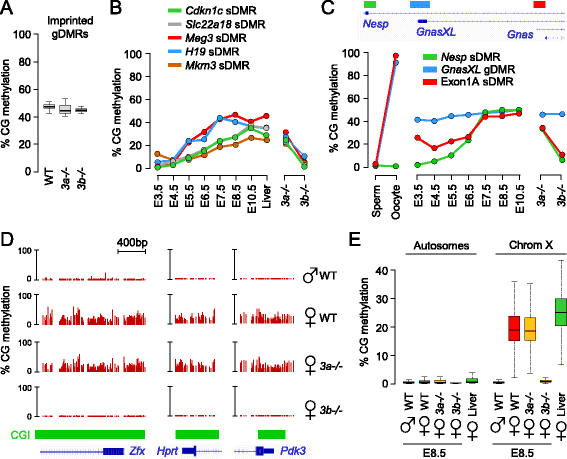
Background: In the mouse, the patterns of DNA methylation are established during early embryonic development in the epiblast. We quantified the targets and kinetics of DNA methylation acquisition in epiblast cells, and determined the contribution of the de novo methyltransferases DNMT3A and DNMT3B to this process.
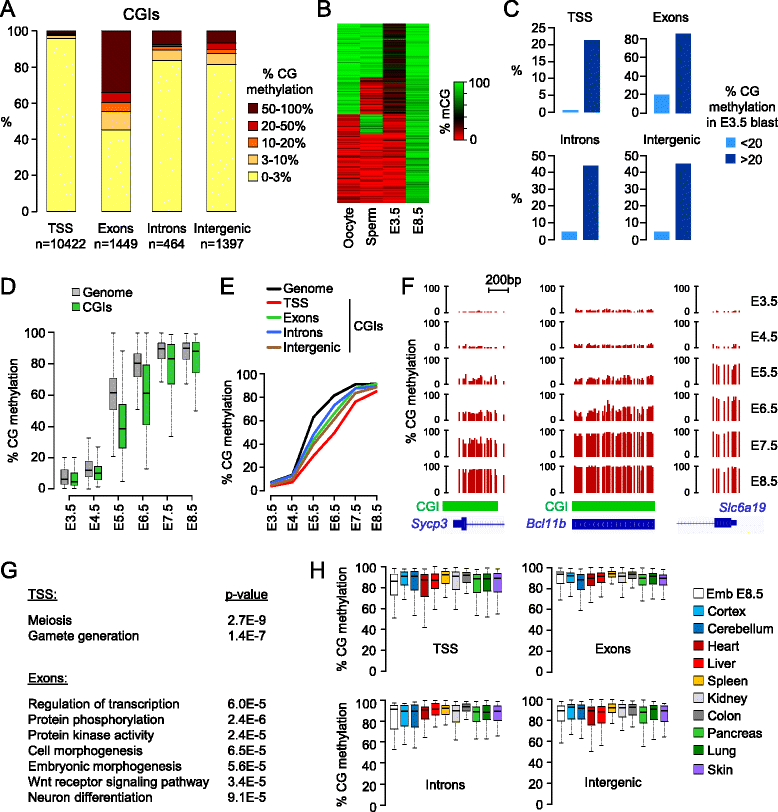
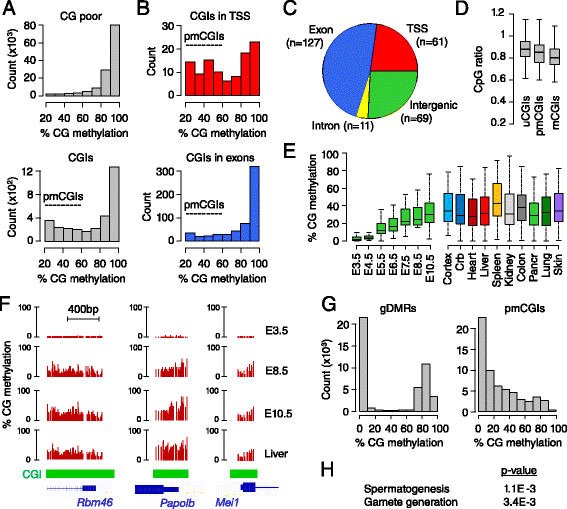
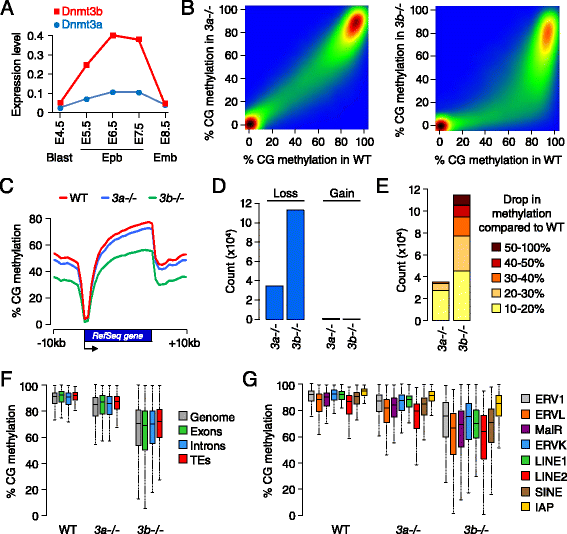
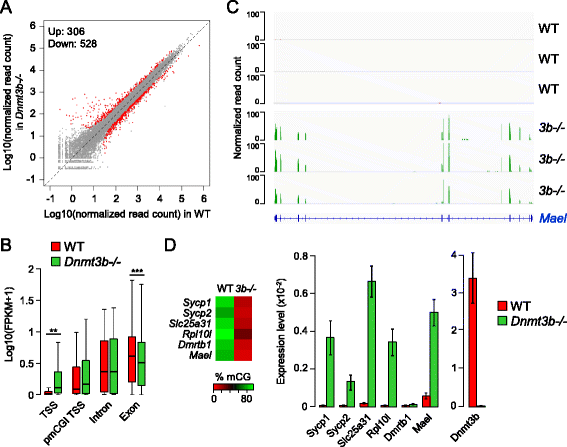
Results: We generated single-base maps of DNA methylation from the blastocyst to post-implantation stages and in embryos lacking DNMT3A or DNMT3B activity. DNA methylation is established within two days of implantation between embryonic days 4.5 and 6.5. The kinetics of de novo methylation are uniform throughout the genome, suggesting a random mechanism of deposition. In contrast, many CpG islands acquire methylation slowly in late epiblast cells. Five percent of CpG islands gain methylation and are found in the promoters of germline genes and in exons of important developmental genes. The onset of global methylation correlates with the upregulation of Dnmt3a/b genes in the early epiblast. DNMT3A and DNMT3B act redundantly to methylate the bulk genome and repetitive elements, whereas DNMT3B has a prominent role in the methylation of CpG islands on autosomes and the X chromosome. Reduced CpG island methylation in Dnmt3b-deficient embryos correlates with gene reactivation in promoters but reduced transcript abundance in gene bodies. Finally, DNMT3B establishes secondary methylation marks at imprinted loci, which distinguishes bona fide germline from somatic methylation imprints.
Conclusions: We reveal that the DNMT3 de novo methyltransferases play both redundant and specific functions in the establishment of DNA methylation in the mouse embryo.
Figures


References
-
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. - PubMed
-
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

