Organization and evolution of transposable elements along the bread wheat chromosome 3B
- PMID: 25476263
- PMCID: PMC4290129
- DOI: 10.1186/s13059-014-0546-4
Organization and evolution of transposable elements along the bread wheat chromosome 3B
Abstract
Background: The 17 Gb bread wheat genome has massively expanded through the proliferation of transposable elements (TEs) and two recent rounds of polyploidization. The assembly of a 774 Mb reference sequence of wheat chromosome 3B provided us with the opportunity to explore the impact of TEs on the complex wheat genome structure and evolution at a resolution and scale not reached so far.
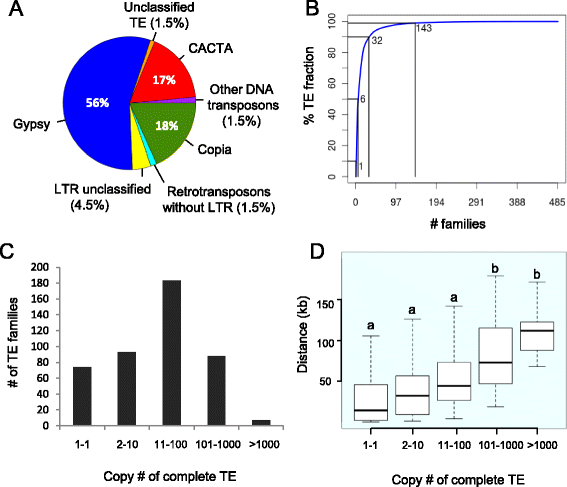
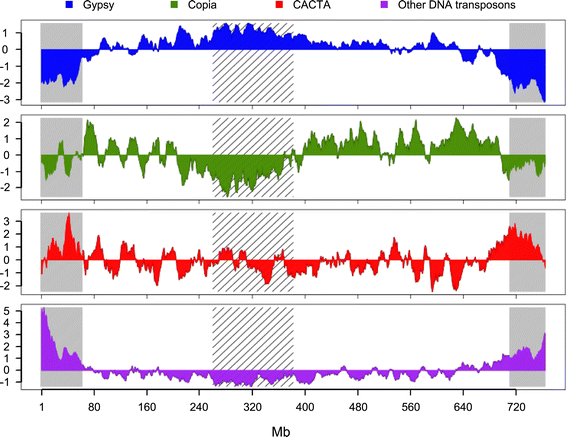
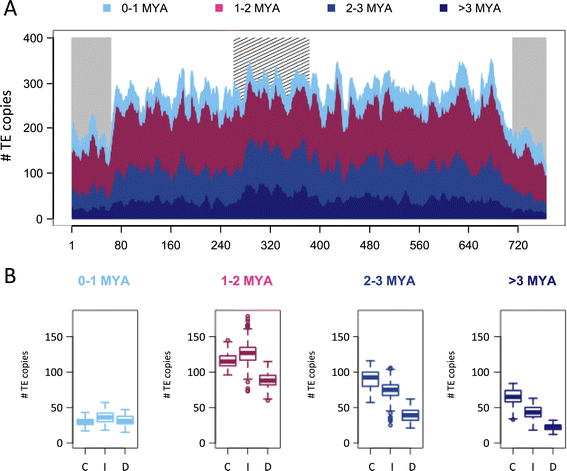
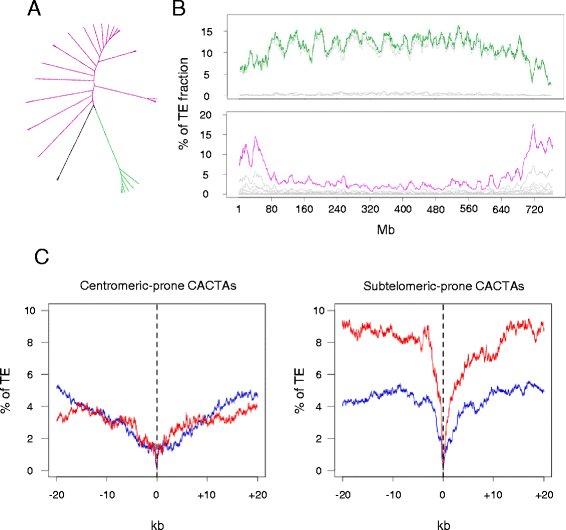
Results: We develop an automated workflow, CLARI-TE, for TE modeling in complex genomes. We delineate precisely 56,488 intact and 196,391 fragmented TEs along the 3B pseudomolecule, accounting for 85% of the sequence, and reconstruct 30,199 nested insertions. TEs have been mostly silent for the last one million years, and the 3B chromosome has been shaped by a succession of bursts that occurred between 1 to 3 million years ago. Accelerated TE elimination in the high-recombination distal regions is a driving force towards chromosome partitioning. CACTAs overrepresented in the high-recombination distal regions are significantly associated with recently duplicated genes. In addition, we identify 140 CACTA-mediated gene capture events with 17 genes potentially created by exon shuffling and show that 19 captured genes are transcribed and under selection pressure, suggesting the important role of CACTAs in the recent wheat adaptation.
Conclusion: Accurate TE modeling uncovers the dynamics of TEs in a highly complex and polyploid genome. It provides novel insights into chromosome partitioning and highlights the role of CACTA transposons in the high level of gene duplication in wheat.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

