DNA methylation and differential gene regulation in photoreceptor cell death
- PMID: 25476906
- PMCID: PMC4649831
- DOI: 10.1038/cddis.2014.512
DNA methylation and differential gene regulation in photoreceptor cell death
Abstract
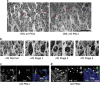
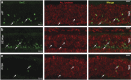
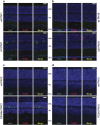
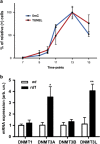
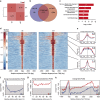
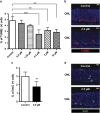
Retinitis pigmentosa (RP) defines a group of inherited degenerative retinal diseases causing progressive loss of photoreceptors. To this day, RP is still untreatable and rational treatment development will require a thorough understanding of the underlying cell death mechanisms. Methylation of the DNA base cytosine by DNA methyltransferases (DNMTs) is an important epigenetic factor regulating gene expression, cell differentiation, cell death, and survival. Previous studies suggested an involvement of epigenetic mechanisms in RP, and in this study, increased cytosine methylation was detected in dying photoreceptors in the rd1, rd2, P23H, and S334ter rodent models for RP. Ultrastructural analysis of photoreceptor nuclear morphology in the rd1 mouse model for RP revealed a severely altered chromatin structure during retinal degeneration that coincided with an increased expression of the DNMT isozyme DNMT3a. To identify disease-specific differentially methylated DNA regions (DMRs) on a genomic level, we immunoprecipitated methylated DNA fragments and subsequently analyzed them with a targeted microarray. Genome-wide comparison of DMRs between rd1 and wild-type retina revealed hypermethylation of genes involved in cell death and survival as well as cell morphology and nervous system development. When correlating DMRs with gene expression data, we found that hypermethylation occurred alongside transcriptional repression. Consistently, motif analysis showed that binding sites of several important transcription factors for retinal physiology were hypermethylated in the mutant model, which also correlated with transcriptional silencing of their respective target genes. Finally, inhibition of DNMTs in rd1 organotypic retinal explants using decitabine resulted in a substantial reduction of photoreceptor cell death, suggesting inhibition of DNA methylation as a potential novel treatment in RP.
Figures

References
-
- 1Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006; 368: 1795–1809. - PubMed
-
- 2Azadi S, Paquet-Durand F, Medstrand P, van Veen T, Ekström PAR. Up-regulation and increased phosphorylation of protein kinase C (PKC) δ, μ and θ in the degenerating rd1 mouse retina. Mol Cell Neurosci 2006; 31: 759–773. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

