Investigation of size-dependent cell adhesion on nanostructured interfaces
- PMID: 25477150
- PMCID: PMC4265325
- DOI: 10.1186/s12951-014-0054-4
Investigation of size-dependent cell adhesion on nanostructured interfaces
Abstract
Background: Cells explore the surfaces of materials through membrane-bound receptors, such as the integrins, and use them to interact with extracellular matrix molecules adsorbed on the substrate surfaces, resulting in the formation of focal adhesions. With recent advances in nanotechnology, biosensors and bioelectronics are being fabricated with ever decreasing feature sizes. The performances of these devices depend on how cells interact with nanostructures on the device surfaces. However, the behavior of cells on nanostructures is not yet fully understood. Here we present a systematic study of cell-nanostructure interaction using polymeric nanopillars with various diameters.
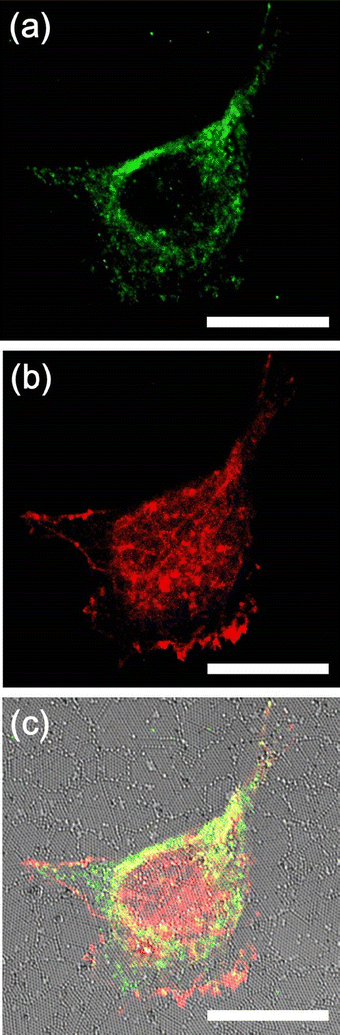
Results: We first checked the viability of cells grown on nanopillars with diameters ranging from 200 nm to 700 nm. It was observed that when cells were cultured on the nanopillars, the apoptosis rate slightly increased as the size of the nanopillar decreased. We then calculated the average size of the focal adhesions and the cell-spreading area for focal adhesions using confocal microscopy. The size of focal adhesions formed on the nanopillars was found to decrease as the size of the nanopillars decreased, resembling the formations of nascent focal complexes. However, when the size of nanopillars decreased to 200 nm, the size of the focal adhesions increased. Further study revealed that cells interacted very strongly with the nanopillars with a diameter of 200 nm and exerted sufficient forces to bend the nanopillars together, resulting in the formation of larger focal adhesions.
Conclusions: We have developed a simple approach to systematically study cell-substrate interactions on physically well-defined substrates using size-tunable polymeric nanopillars. From this study, we conclude that cells can survive on nanostructures with a slight increase in apoptosis rate and that cells interact very strongly with smaller nanostructures. In contrast to previous observations on flat substrates that cells interacted weakly with softer substrates, we observed strong cell-substrate interactions on the softer nanopillars with smaller diameters. Our results indicate that in addition to substrate rigidity, nanostructure dimensions are additional important physical parameters that can be used to regulate behaviour of cells.
Figures
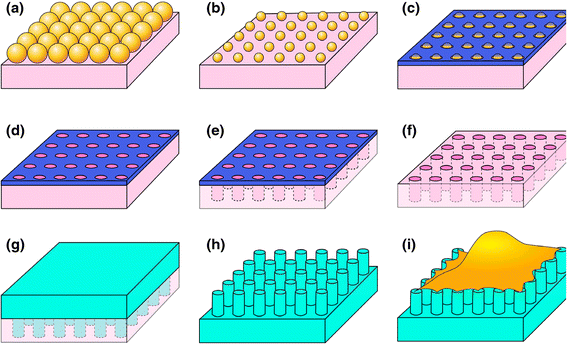
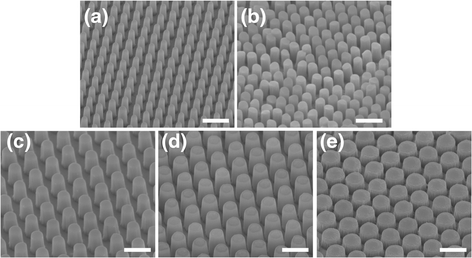

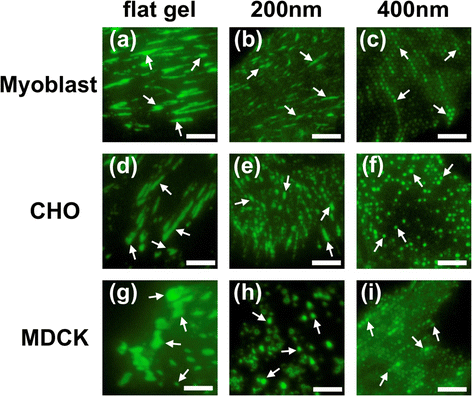

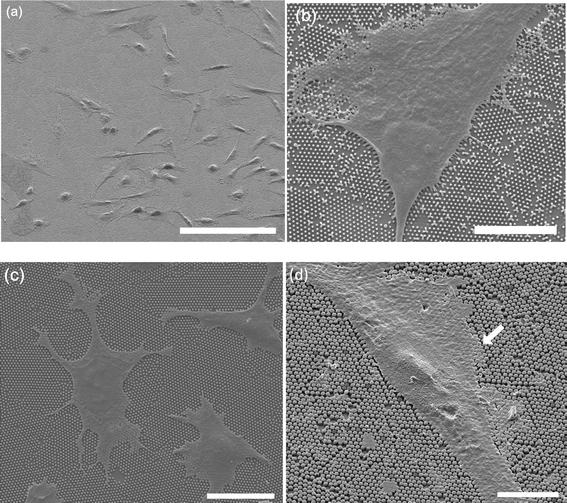

References
-
- Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci. 2001;114:3583–3590. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

