BioAssay Research Database (BARD): chemical biology and probe-development enabled by structured metadata and result types
- PMID: 25477388
- PMCID: PMC4383997
- DOI: 10.1093/nar/gku1244
BioAssay Research Database (BARD): chemical biology and probe-development enabled by structured metadata and result types
Abstract
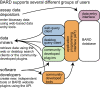
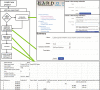
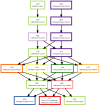
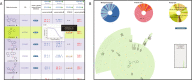
BARD, the BioAssay Research Database (https://bard.nih.gov/) is a public database and suite of tools developed to provide access to bioassay data produced by the NIH Molecular Libraries Program (MLP). Data from 631 MLP projects were migrated to a new structured vocabulary designed to capture bioassay data in a formalized manner, with particular emphasis placed on the description of assay protocols. New data can be submitted to BARD with a user-friendly set of tools that assist in the creation of appropriately formatted datasets and assay definitions. Data published through the BARD application program interface (API) can be accessed by researchers using web-based query tools or a desktop client. Third-party developers wishing to create new tools can use the API to produce stand-alone tools or new plug-ins that can be integrated into BARD. The entire BARD suite of tools therefore supports three classes of researcher: those who wish to publish data, those who wish to mine data for testable hypotheses, and those in the developer community who wish to build tools that leverage this carefully curated chemical biology resource.
Published by Oxford University Press on behalf of Nucleic Acids Research 2014. This work is written by US Government employees and is in the public domain in the US.
Figures

References
-
- Strausberg R.L., Schreiber S.L. From knowing to controlling: a path from genomics to drugs using small molecule probes. Science. 2003;300:294–295. - PubMed
-
- Bolton E.E., Wang Y., Thiessen P.A., Bryant S.H. PubChem: Integrated Platform of Small Molecules and Biological Activities. Vol. 4. Elsevier; 2008. pp. 217–241.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

