A novel RNA molecular signature for activation of 2'-5' oligoadenylate synthetase-1
- PMID: 25477390
- PMCID: PMC4288181
- DOI: 10.1093/nar/gku1289
A novel RNA molecular signature for activation of 2'-5' oligoadenylate synthetase-1
Abstract
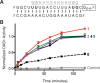
Human 2'-5' oligoadenylate synthetase-1 (OAS1) is central in innate immune system detection of cytoplasmic double-stranded RNA (dsRNA) and promotion of host antiviral responses. However, the molecular signatures that promote OAS1 activation are currently poorly defined. We show that the 3'-end polyuridine sequence of viral and cellular RNA polymerase III non-coding transcripts is critical for their optimal activation of OAS1. Potentiation of OAS1 activity was also observed with a model dsRNA duplex containing an OAS1 activation consensus sequence. We determined that the effect is attributable to a single appended 3'-end residue, is dependent upon its single-stranded nature with strong preference for pyrimidine residues and is mediated by a highly conserved OAS1 residue adjacent to the dsRNA binding surface. These findings represent discovery of a novel signature for OAS1 activation, the 3'-single-stranded pyrimidine (3'-ssPy) motif, with potential functional implications for OAS1 activity in its antiviral and other anti-proliferative roles.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
-
- Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. - PMC - PubMed
-
- Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. - PubMed
-
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

