An ancient defense system eliminates unfit cells from developing tissues during cell competition
- PMID: 25477468
- PMCID: PMC5095928
- DOI: 10.1126/science.1258236
An ancient defense system eliminates unfit cells from developing tissues during cell competition
Abstract
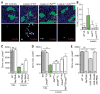
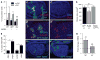
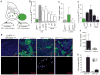
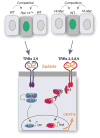
Developing tissues that contain mutant or compromised cells present risks to animal health. Accordingly, the appearance of a population of suboptimal cells in a tissue elicits cellular interactions that prevent their contribution to the adult. Here we report that this quality control process, cell competition, uses specific components of the evolutionarily ancient and conserved innate immune system to eliminate Drosophila cells perceived as unfit. We find that Toll-related receptors (TRRs) and the cytokine Spätzle (Spz) lead to NFκB-dependent apoptosis. Diverse "loser" cells require different TRRs and NFκB factors and activate distinct pro-death genes, implying that the particular response is stipulated by the competitive context. Our findings demonstrate a functional repurposing of components of TRRs and NFκB signaling modules in the surveillance of cell fitness during development.
Copyright © 2014, American Association for the Advancement of Science.
Figures

Comment in
-
Developmental Biology. Death to the losers.Science. 2014 Dec 5;346(6214):1181-2. doi: 10.1126/science.aaa2345. Science. 2014. PMID: 25477441 No abstract available.
References
-
- Buss LW. The Evolution of Individuality. Princeton Univ. Press; Princeton, NJ: 1987.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

