Molecular diagnostics in a teacup: Non-Instrumented Nucleic Acid Amplification (NINA) for rapid, low cost detection of Salmonella enterica
- PMID: 25477717
- PMCID: PMC4251779
- DOI: 10.1007/s11434-012-5634-9
Molecular diagnostics in a teacup: Non-Instrumented Nucleic Acid Amplification (NINA) for rapid, low cost detection of Salmonella enterica
Abstract
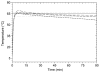
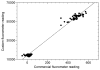
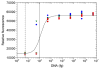
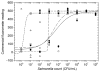
We report on the use of a novel non-instrumented platform to enable a Loop Mediated isothermal Amplification (LAMP) based assay for Salmonella enterica. Heat energy is provided by addition of a small amount (<150 g) of boiling water, and the reaction temperature is regulated by storing latent energy at the melting temperature of a lipid-based engineered phase change material. Endpoint classification of the reaction is achieved without opening the reaction tube by observing the fluorescence of sequence-specific FRET-based assimilating probes with a simple handheld fluorometer. At or above 22°C ambient temperature the non-instrumented devices could maintain reactions above a threshold temperature of 61°C for over 90 min-significantly longer than the 60 min reaction time. Using the simple format, detection limits were less than 20 genome copies for reactions run at ambient temperatures ranging from 8 to 36°C. When used with a pre-enrichment step and non-instrumented DNA extraction device, trace contaminations of Salmonella in milk close to 1 CFU/mL could be reliably detected. These findings illustrate that the non- instrumented amplification approach is a simple, viable, low-cost alternative for field-based food and agricultural diagnostics or clinical applications in developing countries.
Keywords: DNA; LAMP; assimilating probe; biosensor; food safety; molecular diagnostics.
Figures

References
-
- Greig JD, Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. Int J Food Microbiol. 2009;130:77–87. - PubMed
-
- Brooks GF, Butel JS, Carroll KC, et al. Melnick & Adelberg’s Medical Microbiology. New York: McGraw Hill; 2007. Enteric gram-negative rods (enterobacteriaceae)
-
- United States Centers for Disease Control and Prevention. CDC estimates of foodborne illness in the United States. 2011.
-
- Sant’ana AS, Landgraf M, Destro MT, et al. Prevalence and counts of Salmonella spp. In minimally processed vegetables in Sao Paulo, Brazil. Food Microbiol. 2011;28:1235–1237. - PubMed
-
- Kubota R, Jenkins DM, Alvarez AM, et al. Fret-based assimilating probe for sequence specific real-time monitoring of loop mediated isothermal amplification. T Biol Eng. 2011;4:81–100.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
