Microbial sulfur transformations in sediments from Subglacial Lake Whillans
- PMID: 25477865
- PMCID: PMC4237127
- DOI: 10.3389/fmicb.2014.00594
Microbial sulfur transformations in sediments from Subglacial Lake Whillans
Abstract
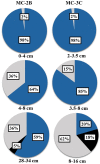
Diverse microbial assemblages inhabit subglacial aquatic environments. While few of these environments have been sampled, data reveal that subglacial organisms gain energy for growth from reduced minerals containing nitrogen, iron, and sulfur. Here we investigate the role of microbially mediated sulfur transformations in sediments from Subglacial Lake Whillans (SLW), Antarctica, by examining key genes involved in dissimilatory sulfur oxidation and reduction. The presence of sulfur transformation genes throughout the top 34 cm of SLW sediments changes with depth. SLW surficial sediments were dominated by genes related to known sulfur-oxidizing chemoautotrophs. Sequences encoding the adenosine-5'-phosphosulfate (APS) reductase gene, involved in both dissimilatory sulfate reduction and sulfur oxidation, were present in all samples and clustered into 16 distinct operational taxonomic units. The majority of APS reductase sequences (74%) clustered with known sulfur oxidizers including those within the "Sideroxydans" and Thiobacillus genera. Reverse-acting dissimilatory sulfite reductase (rDSR) and 16S rRNA gene sequences further support dominance of "Sideroxydans" and Thiobacillus phylotypes in the top 2 cm of SLW sediments. The SLW microbial community has the genetic potential for sulfate reduction which is supported by experimentally measured low rates (1.4 pmol cm(-3)d(-1)) of biologically mediated sulfate reduction and the presence of APS reductase and DSR gene sequences related to Desulfobacteraceae and Desulfotomaculum. Our results also infer the presence of sulfur oxidation, which can be a significant energetic pathway for chemosynthetic biosynthesis in SLW sediments. The water in SLW ultimately flows into the Ross Sea where intermediates from subglacial sulfur transformations can influence the flux of solutes to the Southern Ocean.
Keywords: Antarctic subglacial aquatic environments; chemosynthesis; geomicrobiology; sulfate reduction; sulfur oxidation.
Figures





References
-
- Anderson S. P. (2007). Biogeochemistry of glacial landscape systems. Annu. Rev. Earth Planet. Sci. 35 375–399 10.1126/science.1140766 - DOI
-
- Bak F., Widdel F. (1986). Anaerobic degradation of phenol and phenol derivatives by Desulfobacterium phenolicum sp. nov. Arch. Microbiol. 146:2 177–180 10.1007/BF00402347 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

