D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: implication in vaccine design
- PMID: 25477867
- PMCID: PMC4237132
- DOI: 10.3389/fmicb.2014.00608
D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: implication in vaccine design
Abstract
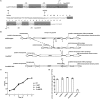
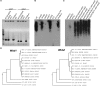
In the O1 strain of Klebsiella, the lipopolysaccharide (LPS) O-antigen is composed of D-galactan I and D-galactan II. Although the composition of the O1 antigen of Klebsiella was resolved more than two decades, the genetic locus involved in the biosynthesis of D-galactan II and the role of D-galactan II in bacterial pathogenesis remain unclear. Here, we report the identification of the D-galactan II-synthesizing genes by screening a transposon mutant library of an acapsulated Klebsiella pneumoniae O1 strain with bacteriophage. K. pneumoniae strain deleted for wbbY exhibited abrogated D-galactan II production; altered serum resistance and attenuation of virulence. Serologic analysis of K. pneumoniae clinical isolates demonstrated that D-galactan II was more prevalent in community-acquired pyogenic liver abscess (PLA)-causing strains than in non-tissue-invasive strains. WbbY homologs, WbbZ homologs, and lipopolysaccharide structures based on D-galactan II also were present in several Gram-negative bacteria. Immunization of mice with the magA-mutant (K(-) 1 O1) (that is, with a LPS D-galactan II-producing strain) provided protection against infection with an O1:K2 PLA strain. Our findings indicate that both WbbY and WbbZ homologs are sufficient for the synthesis of D-galactan II. D-galactan II represents an immunodominant antigen; is conserved among multiple species of Gram-negative bacteria and could be a useful vaccine candidate.
Keywords: D-galactan II; Klebsiella pneumoniae; immunodominant antigen; lipopolysaccharide; vaccine.
Figures



References
-
- Alberti S., Hernandez-Alles S., Gil J., Reina J., Martinez-Beltran J., Camprubi S., et al. . (1993). Development of an enzyme-linked immunosorbent assay method for typing and quantitation of Klebsiella pneumoniae lipopolysaccharide: application to serotype O1. J. Clin. Microbiol. 31, 1379–1381. - PMC - PubMed
-
- Chun K. T., Edenberg H. J., Kelley M. R., Goebl M. G. (1997). Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13, 233–240. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

