LuxR solos in Photorhabdus species
- PMID: 25478328
- PMCID: PMC4235431
- DOI: 10.3389/fcimb.2014.00166
LuxR solos in Photorhabdus species
Abstract
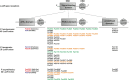
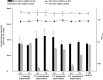
Bacteria communicate via small diffusible molecules to mediate group-coordinated behavior, a process designated as quorum sensing. The basic molecular quorum sensing system of Gram-negative bacteria consists of a LuxI-type autoinducer synthase producing acyl-homoserine lactones (AHLs) as signaling molecules, and a LuxR-type receptor detecting the AHLs to control expression of specific genes. However, many proteobacteria possess one or more unpaired LuxR-type receptors that lack a cognate LuxI-like synthase, referred to as LuxR solos. The enteric and insect pathogenic bacteria of the genus Photorhabdus harbor an extraordinarily high number of LuxR solos, more than any other known bacteria, and all lack a LuxI-like synthase. Here, we focus on the presence and the different types of LuxR solos in the three known Photorhabdus species using bioinformatics analyses. Generally, the N-terminal signal-binding domain (SBD) of LuxR-type receptors sensing AHLs have a motif of six conserved amino acids that is important for binding and specificity of the signaling molecule. However, this motif is altered in the majority of the Photorhabdus-specific LuxR solos, suggesting the use of other signaling molecules than AHLs. Furthermore, all Photorhabdus species contain at least one LuxR solo with an intact AHL-binding motif, which might allow the ability to sense AHLs of other bacteria. Moreover, all three species have high AHL-degrading activity caused by the presence of different AHL-lactonases and AHL-acylases, revealing a high quorum quenching activity against other bacteria. However, the majority of the other LuxR solos in Photorhabdus have a N-terminal so-called PAS4-domain instead of an AHL-binding domain, containing different amino acid motifs than the AHL-sensors, which potentially allows the recognition of a highly variable range of signaling molecules that can be sensed apart from AHLs. These PAS4-LuxR solos are proposed to be involved in host sensing, and therefore in inter-kingdom signaling. Overall, Photorhabdus species are perfect model organisms to study bacterial communication via LuxR solos and their role for a symbiotic and pathogenic life style.
Keywords: LuxR solos; cell-cell communication; entomopathogenic bacteria; quorum quenching; quorum sensing.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

