Temporal and spatial stability of ammonia-oxidizing archaea and bacteria in aquarium biofilters
- PMID: 25479061
- PMCID: PMC4257543
- DOI: 10.1371/journal.pone.0113515
Temporal and spatial stability of ammonia-oxidizing archaea and bacteria in aquarium biofilters
Abstract
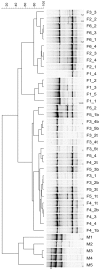
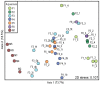
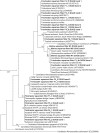
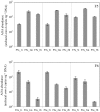
Nitrifying biofilters are used in aquaria and aquaculture systems to prevent accumulation of ammonia by promoting rapid conversion to nitrate via nitrite. Ammonia-oxidizing archaea (AOA), as opposed to ammonia-oxidizing bacteria (AOB), were recently identified as the dominant ammonia oxidizers in most freshwater aquaria. This study investigated biofilms from fixed-bed aquarium biofilters to assess the temporal and spatial dynamics of AOA and AOB abundance and diversity. Over a period of four months, ammonia-oxidizing microorganisms from six freshwater and one marine aquarium were investigated at 4-5 time points. Nitrogen balances for three freshwater aquaria showed that active nitrification by aquarium biofilters accounted for ≥ 81-86% of total nitrogen conversion in the aquaria. Quantitative PCR (qPCR) for bacterial and thaumarchaeal ammonia monooxygenase (amoA) genes demonstrated that AOA were numerically dominant over AOB in all six freshwater aquaria tested, and contributed all detectable amoA genes in three aquarium biofilters. In the marine aquarium, however, AOB outnumbered AOA by three to five orders of magnitude based on amoA gene abundances. A comparison of AOA abundance in three carrier materials (fine sponge, rough sponge and sintered glass or ceramic rings) of two three-media freshwater biofilters revealed preferential growth of AOA on fine sponge. Denaturing gel gradient electrophoresis (DGGE) of thaumarchaeal 16S rRNA genes indicated that community composition within a given biofilter was stable across media types. In addition, DGGE of all aquarium biofilters revealed low AOA diversity, with few bands, which were stable over time. Nonmetric multidimensional scaling (NMDS) based on denaturing gradient gel electrophoresis (DGGE) fingerprints of thaumarchaeal 16S rRNA genes placed freshwater and marine aquaria communities in separate clusters. These results indicate that AOA are the dominant ammonia-oxidizing microorganisms in freshwater aquarium biofilters, and that AOA community composition within a given aquarium is stable over time and across biofilter support material types.
Conflict of interest statement
Figures


References
-
- El-Shafai SA, El-Gohary FA, Nasr FA, van der Steen NP, Gijzen HJ (2004) Chronic ammonia toxicity to duckweed-fed tilapia (Oreochromis niloticus). Aquaculture 232:117–127.
-
- Konneke M, Bernhard AE, de la TorreJR, Walker CB, Waterbury JB, et al. (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. - PubMed
-
- Vlaeminck SE, Hay AG, Maignien L, Verstraete W (2011) In quest of the nitrogen oxidizing prokaryotes of the early Earth. Environ Microbiol 13:283–295. - PubMed
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

