Phytochromes: an atomic perspective on photoactivation and signaling
- PMID: 25480369
- PMCID: PMC4311201
- DOI: 10.1105/tpc.114.131623
Phytochromes: an atomic perspective on photoactivation and signaling
Abstract
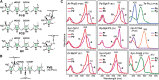
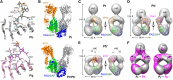
The superfamily of phytochrome (Phy) photoreceptors regulates a wide array of light responses in plants and microorganisms through their unique ability to reversibly switch between stable dark-adapted and photoactivated end states. Whereas the downstream signaling cascades and biological consequences have been described, the initial events that underpin photochemistry of the coupled bilin chromophore and the ensuing conformational changes needed to propagate the light signal are only now being understood. Especially informative has been the rapidly expanding collection of 3D models developed by x-ray crystallographic, NMR, and single-particle electron microscopic methods from a remarkably diverse array of bacterial Phys. These structures have revealed how the modular architecture of these dimeric photoreceptors engages the buried chromophore through distinctive knot, hairpin, and helical spine features. When collectively viewed, these 3D structures reveal complex structural alterations whereby photoisomerization of the bilin drives nanometer-scale movements within the Phy dimer through bilin sliding, hairpin reconfiguration, and spine deformation that ultimately impinge upon the paired signal output domains. When integrated with the recently described structure of the photosensory module from Arabidopsis thaliana PhyB, new opportunities emerge for the rational redesign of plant Phys with novel photochemistries and signaling properties potentially beneficial to agriculture and their exploitation as optogenetic reagents.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures



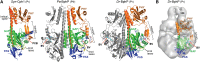



References
-
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. - PubMed
-
- Auldridge M.E., Forest K.T. (2011). Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46: 67–88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

