A major controversy in codon-anticodon adaptation resolved by a new codon usage index
- PMID: 25480780
- PMCID: PMC4317663
- DOI: 10.1534/genetics.114.172106
A major controversy in codon-anticodon adaptation resolved by a new codon usage index
Abstract
Two alternative hypotheses attribute different benefits to codon-anticodon adaptation. The first assumes that protein production is rate limited by both initiation and elongation and that codon-anticodon adaptation would result in higher elongation efficiency and more efficient and accurate protein production, especially for highly expressed genes. The second claims that protein production is rate limited only by initiation efficiency but that improved codon adaptation and, consequently, increased elongation efficiency have the benefit of increasing ribosomal availability for global translation. To test these hypotheses, a recent study engineered a synthetic library of 154 genes, all encoding the same protein but differing in degrees of codon adaptation, to quantify the effect of differential codon adaptation on protein production in Escherichia coli. The surprising conclusion that "codon bias did not correlate with gene expression" and that "translation initiation, not elongation, is rate-limiting for gene expression" contradicts the conclusion reached by many other empirical studies. In this paper, I resolve the contradiction by reanalyzing the data from the 154 sequences. I demonstrate that translation elongation accounts for about 17% of total variation in protein production and that the previous conclusion is due to the use of a codon adaptation index (CAI) that does not account for the mutation bias in characterizing codon adaptation. The effect of translation elongation becomes undetectable only when translation initiation is unrealistically slow. A new index of translation elongation ITE is formulated to facilitate studies on the efficiency and evolution of the translation machinery.
Keywords: codon usage bias; codon-anticodon adaptation; index of translation elongation; translation efficiency; translation elongation.
Copyright © 2015 by the Genetics Society of America.
Figures
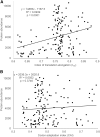

Similar articles
-
Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes.Gene. 2005 Jan 17;345(1):13-20. doi: 10.1016/j.gene.2004.11.019. Epub 2004 Dec 19. Gene. 2005. PMID: 15716092
-
Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization.Genome Res. 2004 Nov;14(11):2279-86. doi: 10.1101/gr.2896904. Epub 2004 Oct 12. Genome Res. 2004. PMID: 15479947 Free PMC article.
-
Conflict between translation initiation and elongation in vertebrate mitochondrial genomes.PLoS One. 2007 Feb 21;2(2):e227. doi: 10.1371/journal.pone.0000227. PLoS One. 2007. PMID: 17311091 Free PMC article.
-
The importance of codon-anticodon interactions in translation elongation.Biochimie. 2015 Jul;114:72-9. doi: 10.1016/j.biochi.2015.04.013. Epub 2015 Apr 25. Biochimie. 2015. PMID: 25921436 Review.
-
Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering.Biotechnol Annu Rev. 2007;13:27-42. doi: 10.1016/S1387-2656(07)13002-7. Biotechnol Annu Rev. 2007. PMID: 17875472 Review.
Cited by
-
Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines.Vaccines (Basel). 2021 Jul 3;9(7):734. doi: 10.3390/vaccines9070734. Vaccines (Basel). 2021. PMID: 34358150 Free PMC article.
-
Quantifying shifts in natural selection on codon usage between protein regions: a population genetics approach.BMC Genomics. 2022 May 30;23(1):408. doi: 10.1186/s12864-022-08635-0. BMC Genomics. 2022. PMID: 35637464 Free PMC article.
-
Elucidating the 16S rRNA 3' boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data.Sci Rep. 2017 Dec 15;7(1):17639. doi: 10.1038/s41598-017-17918-6. Sci Rep. 2017. PMID: 29247194 Free PMC article.
-
Rapid Sampling of Escherichia coli After Changing Oxygen Conditions Reveals Transcriptional Dynamics.Genes (Basel). 2017 Feb 28;8(3):90. doi: 10.3390/genes8030090. Genes (Basel). 2017. PMID: 28264512 Free PMC article.
-
Coevolution mechanisms that adapt viruses to genetic code variations implemented in their hosts.J Genet. 2016 Mar;95(1):3-12. doi: 10.1007/s12041-016-0612-7. J Genet. 2016. PMID: 27019427 No abstract available.
References
-
- Andersson D. I., Kurland C. G., 1983. Ram ribosomes are defective proofreaders. Mol. Gen. Genet. 191: 378–381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

