Oncogenic alterations in ERBB2/HER2 represent potential therapeutic targets across tumors from diverse anatomic sites of origin
- PMID: 25480824
- PMCID: PMC4294606
- DOI: 10.1634/theoncologist.2014-0234
Oncogenic alterations in ERBB2/HER2 represent potential therapeutic targets across tumors from diverse anatomic sites of origin
Abstract
Background: Targeted ERBB2/HER2 inhibitors are approved by the U.S. Food and Drug Administration for the treatment of breast, gastric, and esophageal cancers that overexpress or amplify HER2/ERBB2, as measured by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH), respectively. Activating mutations in ERBB2 have also been reported and are predicted to confer sensitivity to these targeted agents. Testing for these mutations is not performed routinely, and FISH and IHC are not applied outside of these approved indications.
Materials and methods: We explored the spectrum of activating ERBB2 alterations across a collection of ∼ 7,300 solid tumor specimens that underwent comprehensive genomic profiling using next-generation sequencing. Results were analyzed for base substitutions, insertions and deletions, select rearrangements, and copy number changes.
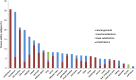
Results: Known oncogenic ERBB2 alterations were identified in tumors derived from 27 tissues, and ERBB2 amplification in breast, gastric, and gastroesophageal cancers accounted for only 30% of these alterations. Activating mutations in ERBB2 were identified in 131 samples (32.5%); amplification was observed in 246 samples (61%). Two samples (0.5%) harbored an ERBB2 rearrangement. Ten samples (2.5%) harbored multiple ERBB2 mutations, yet mutations and amplifications were mutually exclusive in 91% of mutated cases.
Conclusion: Standard slide-based tests for overexpression or amplification of ERBB2 would fail to detect the majority of activating mutations that occur overwhelmingly in the absence of copy number changes. Compared with current clinical standards, comprehensive genomic profiling of a more diverse set of tumor types may identify ∼ 3.5 times the number of patients who may benefit from ERBB2-targeted therapy.
Keywords: Antibodies; High-throughput nucleotide sequencing; Humanized; Lapatinib; Monoclonal; Mutation; Pertuzumab; Trastuzumab.
©AlphaMed Press.
Conflict of interest statement
Disclosures of potential conflicts of interest may be found at the end of this article.
Figures

References
-
- National Research Council (U.S.) Committee on A Framework for Developing a New Taxonomy of Disease. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, D.C.: National Academies Press; 2011. - PubMed
-
- Chmielecki J, Meyerson M. DNA sequencing of cancer: What have we learned? Annu Rev Med. 2014;65:63–79. - PubMed
-
- Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. - PubMed
-
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. - PubMed
-
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

