Epigenetic dysregulation of hairy and enhancer of split 4 (HES4) is associated with striatal degeneration in postmortem Huntington brains
- PMID: 25480889
- PMCID: PMC4321450
- DOI: 10.1093/hmg/ddu561
Epigenetic dysregulation of hairy and enhancer of split 4 (HES4) is associated with striatal degeneration in postmortem Huntington brains
Abstract
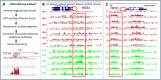
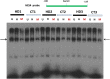
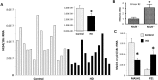
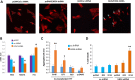
To investigate epigenetic contributions to Huntington's disease (HD) pathogenesis, we carried out genome-wide mapping of the transcriptional mark, trimethyl-histone H3-lysine 4 (H3K4me3) in neuronal nuclei extracted from prefrontal cortex of HD cases and controls using chromatin immunoprecipitation followed by deep-sequencing. Neuron-specific mapping of the genome-wide distribution of H3K4me3 revealed 136 differentially enriched loci associated with genes implicated in neuronal development and neurodegeneration, including GPR3, TMEM106B, PDIA6 and the Notch signaling genes hairy and enhancer of split 4 (HES4) and JAGGED2, supporting the view that the neuronal epigenome is affected in HD. Importantly, loss of H3K4me3 at CpG-rich sequences on the HES4 promoter was associated with excessive DNA methylation, reduced binding of nuclear proteins to the methylated region and altered expression of HES4 and HES4 targeted genes MASH1 and P21 involved in striatal development. Moreover, hypermethylation of HES4 promoter sequences was strikingly correlated with measures of striatal degeneration and age-of-onset in a cohort of 25 HD brains (r = 0.56, P = 0.006). Lastly, shRNA knockdown of HES4 in human neuroblastoma cells altered MASH1 and P21 mRNA expression and markedly increased mutated HTT-induced aggregates and cell death. These findings, taken together, suggest that epigenetic dysregulation of HES4 could play a critical role in modifying HD disease pathogenesis and severity.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Martin J.B., Gusella J.F. Huntington's disease. Pathogenesis and management. N. Engl. J. Med. 1986;315:1267–1276. - PubMed
-
- Vonsattel J.P., DiFiglia M. Huntington disease. J. Neuropathol. Exp. Neurol. 1998;57:369–384. - PubMed
-
- Group, T.H.s.D.C.R. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. - PubMed
-
- MacDonald M.E., Gines S., Gusella J.F., Wheeler V.C. Huntington's disease. Neuromol. Med. 2003;4:7–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

