Human Hematopoietic Signal peptide-containing Secreted 1 (hHSS1) modulates genes and pathways in glioma: implications for the regulation of tumorigenicity and angiogenesis
- PMID: 25481245
- PMCID: PMC4295481
- DOI: 10.1186/1471-2407-14-920
Human Hematopoietic Signal peptide-containing Secreted 1 (hHSS1) modulates genes and pathways in glioma: implications for the regulation of tumorigenicity and angiogenesis
Abstract
Background: Human Hematopoietic Signal peptide-containing Secreted 1 (hHSS1) is a truly novel protein, defining a new class of secreted factors. We have previously reported that ectopic overexpression of hHSS1 has a negative modulatory effect on cell proliferation and tumorigenesis in glioblastoma model systems. Here we have used microarray analysis, screened glioblastoma samples in The Cancer Genome Atlas (TCGA), and studied the effects of hHSS1 on glioma-derived cells and endothelial cells to elucidate the molecular mechanisms underlying the anti-tumorigenic effects of hHSS1.
Methods: Gene expression profiling of human glioma U87 and A172 cells overexpressing hHSS1 was performed. Ingenuity® iReport™ and Ingenuity Pathway Analysis (IPA) were used to analyze the gene expression in the glioma cells. DNA content and cell cycle analysis were performed by FACS, while cell migration, cell invasion, and effects of hHSS1 on HUVEC tube formation were determined by transwell and matrigel assays. Correlation was made between hHSS1 expression and specific genes in glioblastoma samples in the TCGA database.
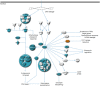
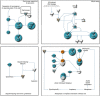
Results: We have clarified the signaling and metabolic pathways (i.e. role of BRCA1 in DNA damage response), networks (i.e. cell cycle) and biological processes (i.e. cell division process of chromosomes) that result from hHSS1effects upon glioblastoma growth. U87-overexpressing hHSS1 significantly decreased the number of cells in the G0/G1 cell cycle phase, and significantly increased cells in the S and G2/M phases (P < 0.05). U87-overexpressing hHSS1 significantly lost their ability to migrate (P < 0.001) and to invade (P < 0.01) through matrigel matrix. hHSS1-overexpression significantly decreased migration of A172 cells (P < 0.001), inhibited A172 tumor-induced migration and invasion of HUVECs (P < 0.001), and significantly inhibited U87 tumor-induced invasion of HUVECs (P < 0.001). Purified hHSS1 protein inhibited HUVEC tube formation. TCGA database revealed significant correlation between hHSS1 and BRCA2 (r = -0.224, P < 0.0005), ADAMTS1 (r = -0.132, P <0.01) and endostatin (r = 0.141, P < 0.005).
Conclusions: hHSS1-overexpression modulates signaling pathways involved in tumorigenesis. hHSS1 inhibits glioma-induced cell cycle progression, cell migration, invasion and angiogenesis. Our data suggest that hHSS1 is a potential therapeutic for malignant glioblastoma possessing significant antitumor and anti-angiogenic activity.
Figures







References
-
- Moore K, Kim L. Primary Brain Tumors: Characteristics, Pratical Diagnostic and Treatment Approaches. In: Ray SK, editor. Glioblastoma: Molecular Mechanisms of Pathogenesis and Current Therapeutic Strategies. 2010. pp. 43–75.
Pre-publication history
-
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/920/prepub
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

