Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1
- PMID: 25481746
- PMCID: PMC4381436
- DOI: 10.1016/j.jmb.2014.11.018
Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1
Abstract
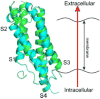
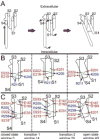
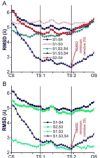
Voltage-gated proton channels (Hv1) are ubiquitous throughout nature and are implicated in numerous physiological processes. The gene encoding for Hv1, however, was only identified in 2006. The lack of sufficient structural information of this channel has hampered the understanding of the molecular mechanism of channel activation and proton permeation. This study uses both simulation and experimental approaches to further develop existing models of the Hv1 channel. Our study provides insights into features of channel gating and proton permeation pathway. We compare open- and closed-state structures developed previously with a recent crystal structure that traps the channel in a presumably closed state. Insights into gating pathways were provided using a combination of all-atom molecular dynamics simulations with a swarm of trajectories with the string method for extensive transition path sampling and evolution. A detailed residue-residue interaction profile and a hydration profile were studied to map the gating pathway in this channel. In particular, it allows us to identify potential intermediate states and compare them to the experimentally observed crystal structure of Takeshita et al. (Takeshita K, Sakata S, Yamashita E, Fujiwara Y, Kawanabe A, Kurokawa T, et al. X-ray crystal structure of voltage-gated proton channel. Nature 2014). The mechanisms governing ion transport in the wild-type and mutant Hv1 channels were studied by a combination of electrophysiological recordings and free energy simulations. With these results, we were able to further refine ideas about the location and function of the selectivity filter. The refined structural models will be essential for future investigations of this channel and the development of new drugs targeting cellular proton transport.
Keywords: gating mechanism; ion transport; voltage-gated proton channels.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures






References
-
- Takeshita K, Sakata S, Yamashita E, Fujiwara Y, Kawanabe A, Kurokawa T, et al. X-ray crystal structure of voltage-gated proton channel. Nature. 2014 - PubMed
-
- Thomas RC, Meech RW. Hydrogen-Ion Currents and Intracellular Ph in Depolarized Voltage-Clamped Snail Neurons. Nature. 1982;299:826–8. - PubMed
-
- Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

