The structure function of the death domain of human IRAK-M
- PMID: 25481771
- PMCID: PMC4273448
- DOI: 10.1186/s12964-014-0077-3
The structure function of the death domain of human IRAK-M
Abstract
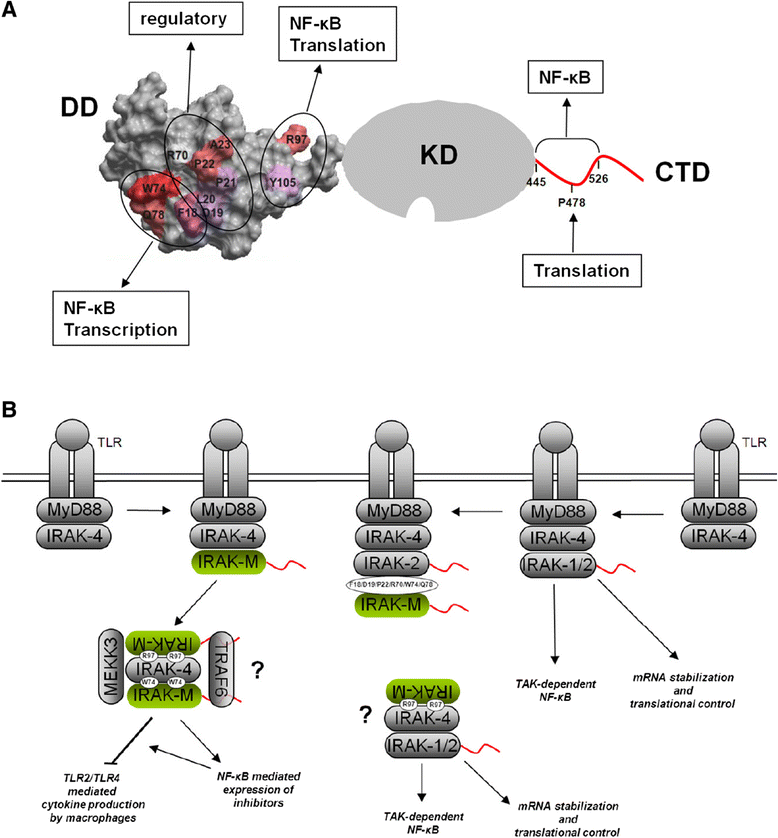
Background: IRAK-M is an inhibitor of Toll-like receptor signaling that acts by re-directing IRAK-4 activity to TAK1 independent NF-κB activation and by inhibition of IRAK-1/IRAK-2 activity. IRAK-M is expressed in monocytes/macrophages and lung epithelial cells. Lack of IRAK-M in mice greatly improves the resistance to nosocomial pneumonia and lung tumors, which entices IRAK-M as a potential therapeutic target. IRAK-M consists of an N-terminal death domain (DD), a dysfunctional kinase domain and unstructured C-terminal domain. Little is known however on IRAK-M's structure-function relationships.
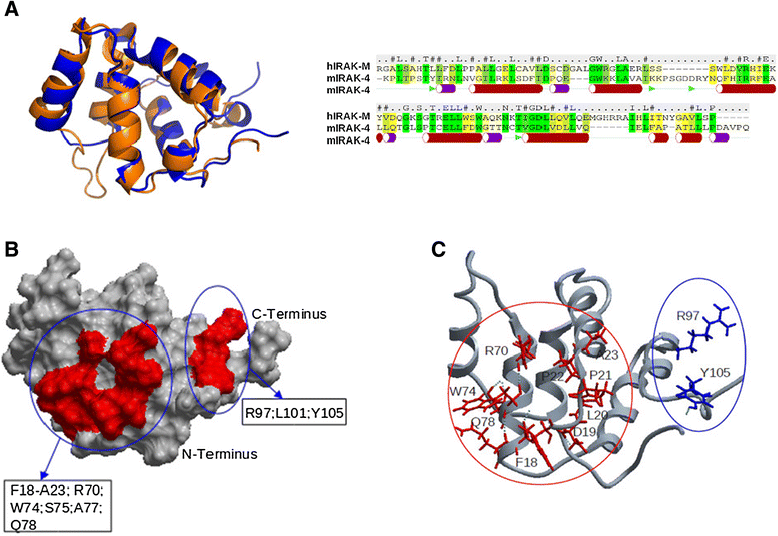
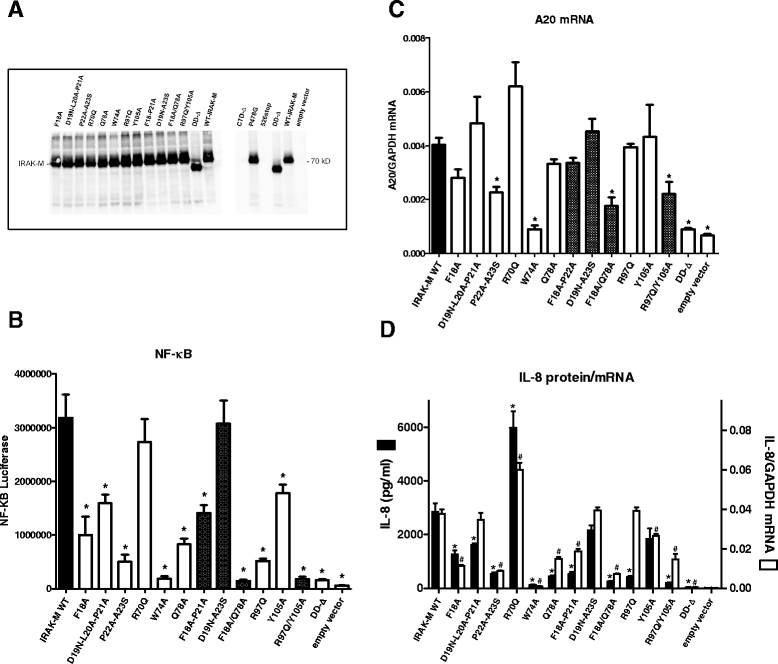
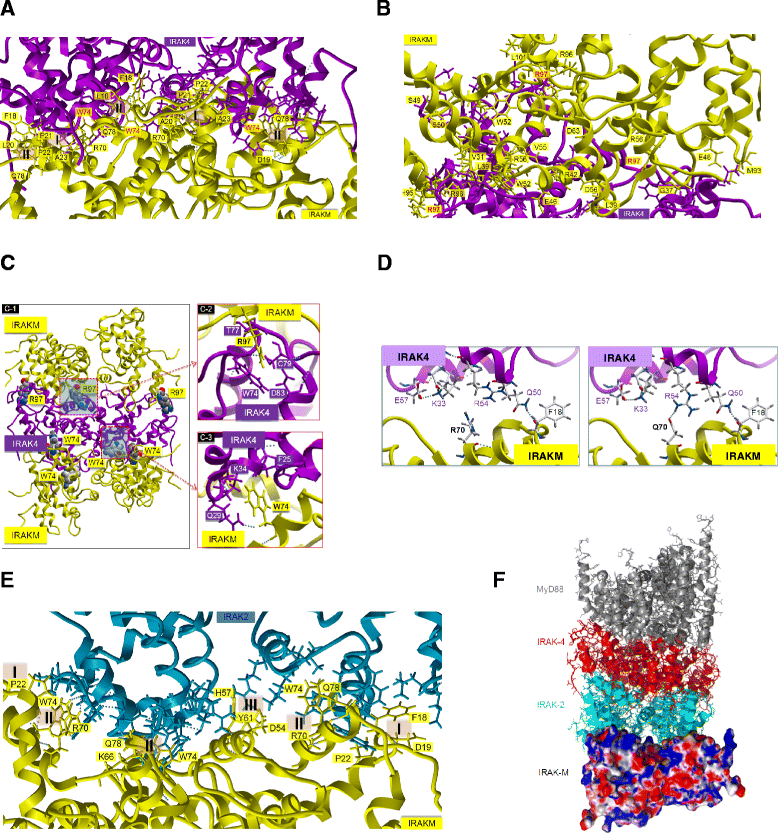
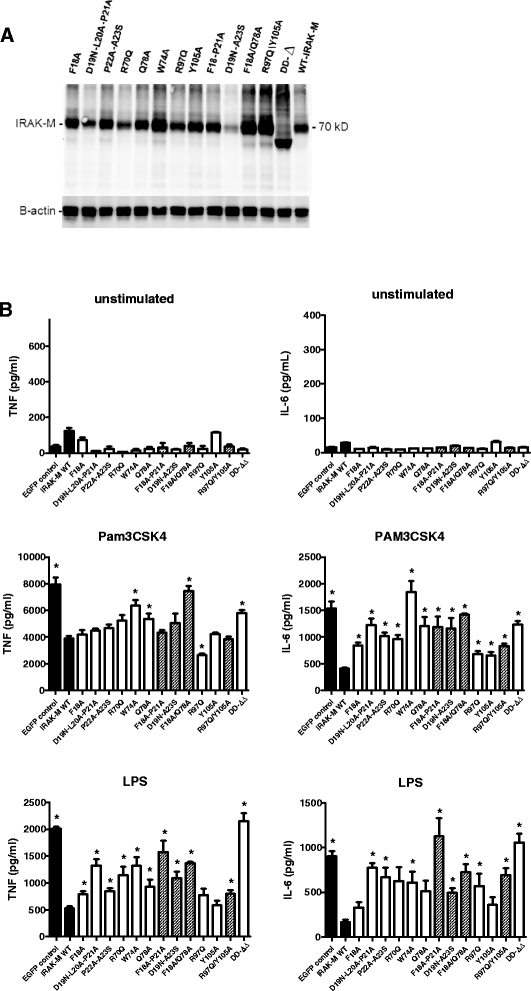
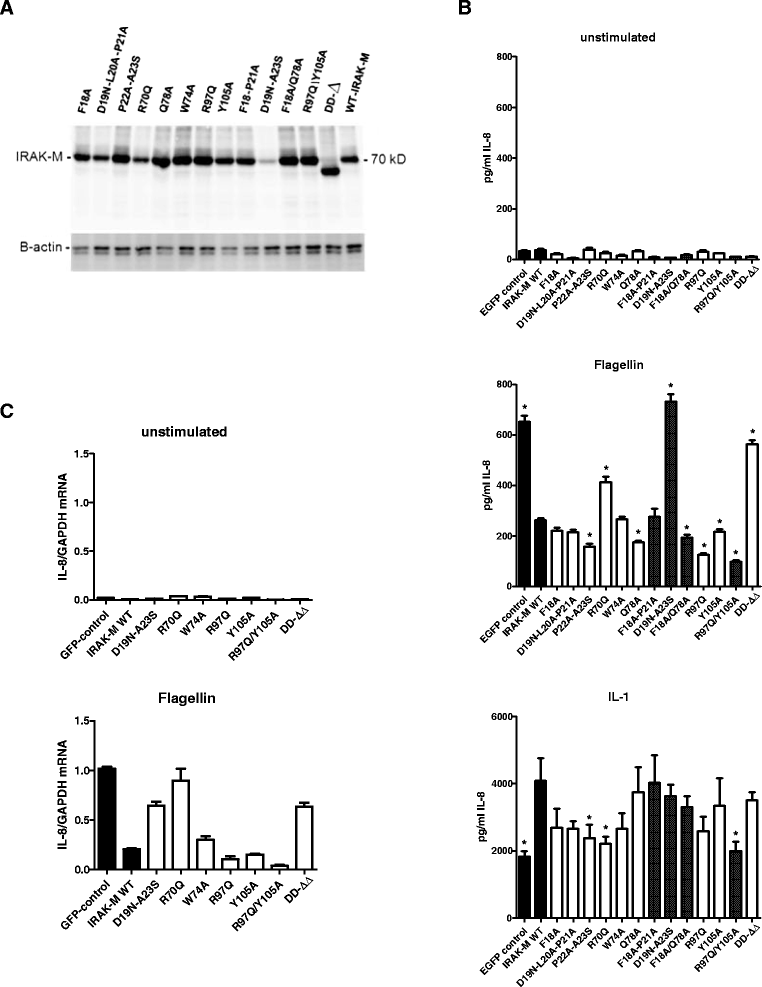
Results: Since death domains provide the important interactions of IRAK-1, IRAK-2 and IRAK-4 molecules, we generated a 3D structure model of the human IRAK-M-DD (residues C5-G119) to guide mutagenesis studies and predict protein-protein interaction points. First we identified the DD residues involved in the endogenous capacity of IRAK-M to activate NF-κB that is displayed upon overexpression in 293T cells. W74 and R97, at distinct interfaces of the IRAK-M-DD, were crucial for this endogenous NF-κB activating capacity, as well as the C-terminal domain (S445-E596) of IRAK-M. Resulting anti-inflammatory A20 and pro-inflammatory IL-8 transcription in 293T cells was W74 dependent, while IL-8 protein expression was dependent on R97 and the TRAF6 binding motif at P478. The IRAK-M-DD W74 and R97 binding interfaces are predicted to interact with opposite sides of IRAK-4-DD's. Secondly we identified DD residues important for the inhibitory action of IRAK-M by stable overexpression of mutants in THP-1 macrophages and H292 lung epithelial cells. IRAK-M inhibited TLR2/4-mediated cytokine production in macrophages in a manner that is largely dependent on W74. R97 was not involved in inhibition of TNF production but was engaged in IL-6 down-regulation by IRAK-M. Protein-interactive residues D19-A23, located in between W74 and R97, were also observed to be crucial for inhibition of TLR2/4 mediated cytokine induction in macrophages. Remarkably, IRAK-M inhibited TLR5 mediated IL-8 production by lung epithelial cells independent of W74 and R97, but dependent on D19-A23 and R70, two surface-exposed regions that harbor predicted IRAK-2-DD interaction points of IRAK-M.
Conclusion: IRAK-M employs alternate residues of its DD to inhibit the different inflammatory mediators induced by varying TLRs and cells.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

