PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model
- PMID: 25482129
- PMCID: PMC4456291
- DOI: 10.1038/leu.2014.341
PTPROt-mediated regulation of p53/Foxm1 suppresses leukemic phenotype in a CLL mouse model
Abstract
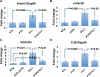
The gene encoding PTPROt (truncated isoform of protein tyrosine phosphatase receptor-type O) is methylated and suppressed in chronic lymphocytic leukemia (CLL). PTPROt exhibits in vitro tumor-suppressor characteristics through the regulation of B-cell receptor (BCR) signaling. Here we generated transgenic (Tg) mice with B-cell-specific expression of PTPROt. Although lymphocyte development is normal in these mice, crossing them with TCL1 Tg mouse model of CLL results in a survival advantage compared with the TCL1 Tg mice. Gene expression profiling of splenic B-lymphocytes before detectable signs of CLL followed by Ingenuity Pathway Analysis revealed that the most prominently regulated functions in TCL1 Tg vs non-transgenic (NTg) and TCL1 Tg vs PTPROt/TCL1 double Tg are the same and also biologically relevant to this study. Further, enhanced expression of the chemokine Ccl3, the oncogenic transcription factor Foxm1 and its targets in TCL1 Tg mice were significantly suppressed in the double Tg mice, suggesting a protective function of PTPROt against leukemogenesis. This study also showed that PTPROt-mediated regulation of Foxm1 involves activation of p53, a transcriptional repressor of Foxm1, which is facilitated through suppression of BCR signaling. These results establish the in vivo tumor-suppressive function of PTPROt and identify p53/Foxm1 axis as a key downstream effect of PTPROt-mediated suppression of BCR signaling.
Figures






References
-
- Wiggins RC, Wiggins JE, Goyal M, Wharram BL, Thomas PE. Molecular cloning of cDNAs encoding human GLEPP1, a membrane protein tyrosine phosphatase: characterization of the GLEPP1 protein distribution in human kidney and assignment of the GLEPP1 gene to human chromosome 12p12-p13. Genomics. 1995 May 1;27(1):174–181. - PubMed
-
- Beltran PJ, Bixby JL, Masters BA. Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. The Journal of comparative neurology. 2003 Feb 17;456(4):384–395. - PubMed
-
- Aguiar RC, Yakushijin Y, Kharbanda S, Tiwari S, Freeman GJ, Shipp MA. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood. 1999 Oct 1;94(7):2403–2413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

